After preprocessing with fMRIprep, there are minor differences in the orientation information of functional images, which, although small, lead to errors when using SPM for further analysis.
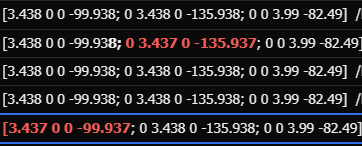
(This is part of the orientation information)
I’m puzzled , as the portion with differences accounts for only 16% of the total images, but I used the same command to process all the images.
We will not be able to help with the information provided. I’ve moved this post to the Software Support category and copied in the default template. Please edit your post and fill in all of the information you can.
Maybe someone else can comment on why these minor differences in the last decimal place happened exactly, but it’s likely some difference in rounding or reading and writing from a text file somewhere along the processing. In any case, these are possible workarounds that change the header of the datasets to match each other. Mucking around with the header is risky, so work with copies of the data.
- nifti_tool. Use the “-mod_hdr -mod_field” options to change fields, copying the values from one of the datasets to the others. Check the fields with “nifti_tool -disp_hdr -infiles …”. This extract shows the ones that deal with orientation and voxel dimensions:
pixdim 76 8 1.0 0.25 0.25 0.25 0.0 0.0 0.0 0.0
qform_code 252 1 5
sform_code 254 1 5
quatern_b 256 1 0.0
quatern_c 260 1 -0.0
quatern_d 264 1 0.0
qoffset_x 268 1 -39.75
qoffset_y 272 1 -58.0
qoffset_z 276 1 -32.25
srow_x 280 4 0.25 -0.0 -0.0 -39.75
srow_y 296 4 -0.0 0.25 -0.0 -58.0
srow_z 312 4 0.0 0.0 0.25 -32.25
- 3drefit. AFNI’s 3drefit can copy over attributes from one dataset to another. This should work with multiple NIFTI files as input:
3drefit -atrcopy IJK_TO_DICOM_REAL original_dset modified_dsets
- SPM matlab. Since you are working with SPM, you might consider the Matlab way proposed here to copy headers.
matlab - Copy one Nifti header to another - Stack Overflow
hi@dglen,
Thanks so much,I truly believe the various methods you provided can fully address my Issue!