Hello,
I’m happy to use BIDSCOIN v4.3 to convert a dataset using PAR/REC format, which I’m not used to.
I face several issues and I would be happy to get some inputs because I’m stucked at this point 
A) Several scans have the same parameters (except the Acquisition number) but I fail to separate them using a tag (only <>).
This is an issue for this protocol since the 2 repeated scans have to be tagged “preTraining” and “PostTraining” due to an habituation effect in the scanner.
I precise that they were collected in the same MRI session, without exit.
Is there another way to tag these 2 “same” scans, please?
B) I’m also struggling with 2 field maps scans.
I’m so used to Siemens fieldmaps scans (with 2 magnitude & 1 phase difference scan that I’m lost).
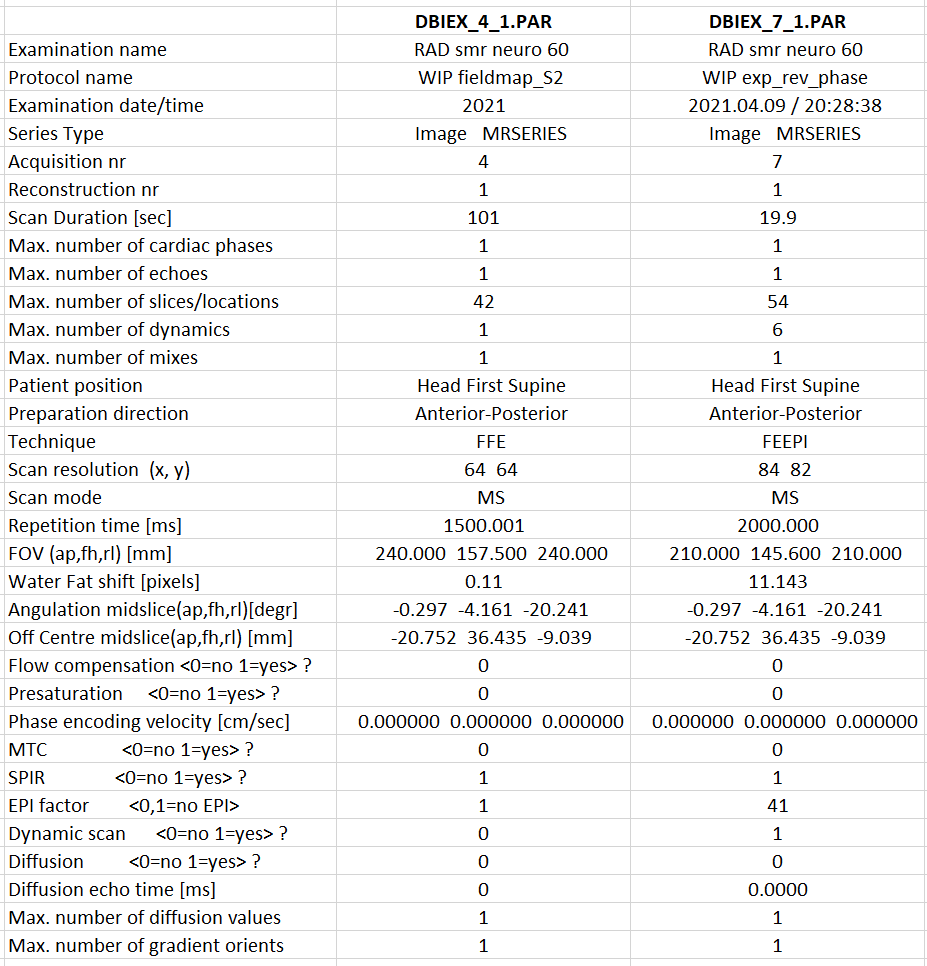
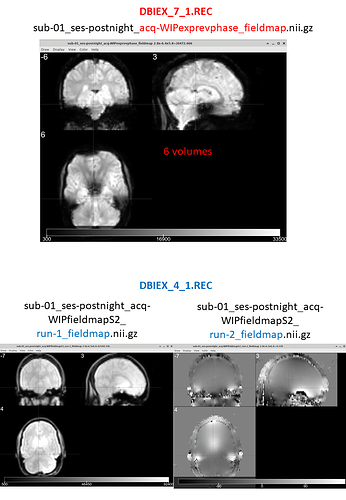
Here are the headers & images of the 2 PAR files:
In the BIDSeditor, I chose the “fieldmap” suffix and I get:
2 nifti volumes (run-1 & run-2) from the DBIEX_4_1.PAR scan (which look like to me as 1 magnitude & 1 phase scans)
a 4D nifti volume from the DBIEX_7_1.PAR scan (which look like EPI scans)
Does it make sense to label these scans as fieldmaps?
Are they usable for the preprocessing of the BOLD scans? (I’m used to fmriprep using the IntendedFor option in the json files).
C) For each of the 30 participants, 2 scans were interrupted according their response time in the scanner.
This was planned from the beginning to let them enough time to answer, which was not predictable.
However, for each scan, this causes a (fatal) ERROR by BIDSCOIN since “Error: Total number of slices (5738) not divisible by slices per 3D volume (54) [acquisition aborted].”
The advised tool, namely, R2AGUI works fine but I would like to know if there is an option/a trick to make it work using BIDSCOIN (this would save some time for me to process these scans).
That would be the cherry but I can deal with that, if not possible.
(and if I may, I would love to add the phys2bidscoin option available in my bidseditor but I never managed ; phys2bidscoin works fine though outside the gui interface like in Options — BIDScoin 4.3.2+qt5 documentation).
Thank you for any help or suggestion!
Best,
Gaëlle
A) So if they don’t differ in their header attributes, do they then perhaps differ in their file properties? BIDScoin can also separate runs (using <>) using (part of) their filename or filepath. If not, there is no information to work with, except that run-1 needs to be tagged as “preTraining” and run-2 as “postTraining”. You can of course rename your files as a post-processing procedure, but this is not optimal. What you also can do, before running the bidsmapper, is “extend” the PAR/REC attributes by adding (very simple) json sidecar files to the files you would like to separate. For instance, the pre-training json files could look something like this:
{"protocol_name": "preTraining"}
and your post-training jsons like this:
{"protocol_name": "postTraining"}
Then you in your bidsmap you can use e.g. acq: <protocol_name>
B) I am also not used to PAR/REC at all, and I don’t know much about how Philips stores fieldmaps. Perhaps someone here can explain what is what, then I can help you to properly map it all to BIDS
C) This error you posted about the number of slices is not from BIDScoin, but is generated by dcm2niix. Dcm2niix supports PAR/REC, but it is not the best PAR/REC converter (I believe the older dcm2nii is even supposed to do a better job). BIDScoin doesn’t have a plugin for R2AGUI, but you could try installing dcm2nii and use it as a drop-in replacement for dcm2niix. AFter installing it, all you have to do is replace dcm2niix with dcm2nii in your bidsmap options.
As for phys2bidscoin, that was a hackathon project that was never finished. You said: " phys2bidscoin works fine though outside the gui interface". That I don’t understand. Do you mean that phys2bids itself works fine on your data? Because you can always run that separately.
Hope this helps
For PAR/REC data, I would recommend dicm2nii - it requires Matlab but is a terrific tool. Regardless of what tool you use, be aware that the raw PAR/REC files are missing many of the details required to conduct reproducible neuroscience, so the BIDS validator will detect missing fields. This is not a limitation of the converter used, rather it reflects the impoverished inputs. Where possible, I would recommend exporting to DICOM rather than PAR/REC.
I would urge all users of Philips scanners to lobby the Philips Clinical Scientists associated with their site to update their software to store required sequence details in the DICOM images. Philips makes terrific software, but users will have to provide sequence details manually until this is resolved.
Hello,
Thanks a lot for your help, both of you ; it’s really useful for me.
Indeed, I’ll go for dicm2nii since I can access Matlab here. After testing it on the files to be converted, it helped to disentangle the fielmap scan N°4, creating a pair of files with *_real.nii.gz and *_magnitude.nii.gz
It also helps to deal with the scan N°8 with an interrupted data collection.
I’ll go for this, thanks a lot.
(I’ll forward your message about the source data format. Hopefully, it will help to switch to dicom!).
Best, Gaëlle
Sounds good. What you can do after running dicm2nii is to use BIDScoin’s nibabel plugin to easily “convert” (reorganise) your nifti’s into BIDS
Thanks for your advice. it works fine and helps a lot, again!
Just a last issue with the fmaps to fix and it would be good.
Best, Gaëlle
![]()