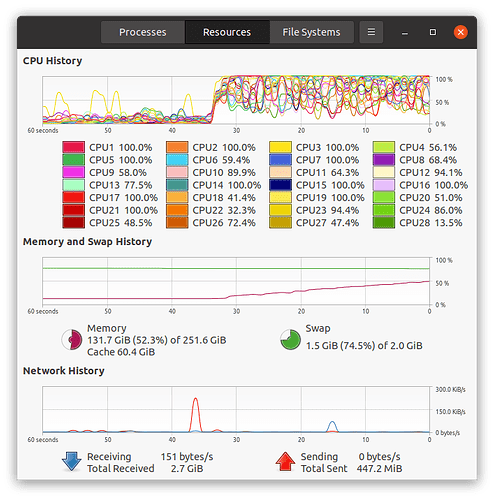
Dear @bthirion , thanks again for your help.
The solution was indeed to provide a fix value to n_jobs_cv and remove the del statement, but also to completely restructure the script, because paralleling had to be called at the beginning of each participants, not within the loop.
Great succes!
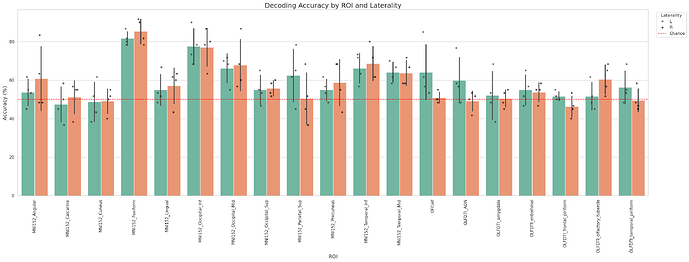
The results for 4 subjects, 18 ROI (L&R), 6 runs, 18 trials (140 images, 2x2x2 3T), leave 2 runs out are obtained in ~26 minutes.
For those interested, please find my (quite long) script bellow. In my case, I am looking at the difference in accuracy scores between 2 conditions (and so, types of runs, v and vo).
import pandas as pd
import os
nilearn_folder = "nilearn_se"
current_directory = os.getcwd()
source_data_directory = os.path.abspath(os.path.join(current_directory, "..", nilearn_folder, "sourcedata"))
run_and_trial_types = pd.read_csv(os.path.join(source_data_directory, "run_and_trial_types.csv"))
run_and_trial_types.drop(["Unnamed: 0"], axis=1, inplace=True)
run_and_trial_types
import os
import re
import pandas as pd
import numpy as np
import time
from datetime import datetime
import matplotlib.pyplot as plt
from nilearn.image import load_img, mean_img
from nilearn.plotting import plot_epi
from nilearn.glm.first_level import FirstLevelModel
from nilearn.decoding import Decoder
from nilearn.reporting import make_glm_report
from nilearn.maskers import NiftiMasker
from sklearn.model_selection import LeaveOneGroupOut
import warnings
warnings.filterwarnings("ignore", category=UserWarning)
#------------------Parameters------------------
data_location = "nilearn_se"
task_name = "odor"
t_r = 1.5
n_runs = 12
run_type="vo"
n_runs_to_train_test = 6
regroup_by_domain = True # regroup by domain (ex: images 2,3 and 4 -> Food)
domain_mapping = {
'img/obj2': 'Food',
'img/obj3': 'Food',
'img/obj4': 'Food',
'img/obj5': 'Human',
'img/obj6': 'Human',
'img/obj7': 'Human',
}
subject_list = [
"01", "02", "03", "04"
]
roi_names = [
# VISION
# "MNI152_visual_all_cortex_LR", "MNI152_visual_all_cortex_L", "MNI152_visual_all_cortex_R", # created by combining all VC L & R ROI
# "MNI152_visual_primary_cortex_LR", "MNI152_visual_primary_cortex_L", "MNI152_visual_primary_cortex_R",
# "MNI152_visual_secondary_cortex_LR", "MNI152_visual_secondary_cortex_L", "MNI152_visual_secondary_cortex_R",
# "MNI152_visual_ventral_cortex_LR", "MNI152_visual_ventral_cortex_L", "MNI152_visual_ventral_cortex_R",
# "MNI152_visual_dorsal_cortex_LR", "MNI152_visual_dorsal_cortex_L", "MNI152_visual_dorsal_cortex_R",
"MNI152_Lingual_L_1mm", "MNI152_Lingual_R_1mm",
"MNI152_Cuneus_L_1mm", "MNI152_Cuneus_R_1mm",
"MNI152_Calcarine_L_1mm", "MNI152_Calcarine_R_1mm",
"MNI152_Occipital_Sup_L_1mm", "MNI152_Occipital_Sup_R_1mm",
"MNI152_Occipital_Mid_L_1mm", "MNI152_Occipital_Mid_R_1mm",
"MNI152_Occipital_Inf_L_1mm", "MNI152_Occipital_Inf_R_1mm",
"MNI152_Fusiform_L_1mm", "MNI152_Fusiform_R_1mm",
"MNI152_Temporal_Inf_L_1mm", "MNI152_Temporal_Inf_R_1mm",
"MNI152_Temporal_Mid_L_1mm", "MNI152_Temporal_Mid_R_1mm",
"MNI152_Parietal_Sup_L_1mm", "MNI152_Parietal_Sup_R_1mm",
"MNI152_Precuneus_L_1mm", "MNI152_Precuneus_R_1mm",
"MNI152_Angular_L_1mm", "MNI152_Angular_R_1mm",
# OLFACTION
# "MNI152_olfactory_cortex_LR", "MNI152_olfactory_cortex_L", "MNI152_olfactory_cortex_R", # created by combining all OC L & R ROI
"OLFDTI_R_frontal_piriform", "OLFDTI_L_frontal_piriform",
"OLFDTI_R_temporal_piriform_", "OLFDTI_L_temporal_piriform_",
"OLFDTI_R_amygdala_", "OLFDTI_L_amygdala_",
"OLFDTI_R_AON_", "OLFDTI_L_AON_",
"OLFDTI_R_entorhinal_", "OLFDTI_L_entorhinal_",
"OLFDTI_R_olfactory_tubercle_", "OLFDTI_L_olfactory_tubercle_",
"OFColf_R", "OFColf_L"
]
debug = True
mri_trigger = True # onset = 0 for trial_type == MRI_trigger (realigne t0)
save_stat_maps = True # heavy, will create a stat map for each sub-run-roi-condition
save_glm_report = True # heavy, will create a stat map for each sub-run-roi-condition
save_weight_maps = True # heavy, will create a stat map for each sub-roi-condition
save_confusion_matrix = True
save_final_results = True
#------------------Parameters------------------
# paths
subjfolder = datetime.today().strftime("%Y%m%d")
current_directory = os.getcwd() # Current working directory
bids_dir = os.path.abspath(os.path.join(current_directory, '..', f"{data_location}", 'bids'))
fmriprep_dir = os.path.abspath(os.path.join(current_directory, '..', f"{data_location}", 'fmriprep'))
roi_dir = os.path.abspath(os.path.join(current_directory, '..', f"{data_location}", 'anatomy', 'mni'))
output_dir = os.path.abspath(os.path.join(current_directory, '..', "results", "mvpa", run_type, subjfolder, data_location))
def get_subject_runs(df, run_type="v"):
filt = df["run_type"].str.lower() == run_type.lower()
tmp = df.loc[filt, ["subject_id", "run_id"]].drop_duplicates()
tmp["sub_num"] = tmp["subject_id"].str.extract(r"(\d+)").astype(str)
tmp["run_num"] = tmp["run_id"].str.extract(r"(\d+)").astype(int)
return (
tmp.groupby("sub_num")["run_num"]
.apply(lambda s: sorted(s.tolist()))
.to_dict()
)
def build_unique_runs_per_subject(
df,
run_type=run_type,
n_runs_to_train_test=None
):
runs_by_subject = get_subject_runs(df, run_type)
if n_runs_to_train_test is not None:
runs_by_subject = {
sub: runs[:n_runs_to_train_test]
for sub, runs in runs_by_subject.items()
}
return runs_by_subject
unique_runs = build_unique_runs_per_subject(
run_and_trial_types,
run_type=run_type,
n_runs_to_train_test=n_runs_to_train_test
)
# check prints
print(
f"Data: {data_location}\n"
f"Task: {task_name}\n"
f"Subjects: {subject_list}\n"
f"ROI: {roi_names}\n"
f"TR: {t_r} seconds\n"
f"Number of runs: {len(unique_runs)}\n"
f"Unique runs: {unique_runs}\n"
f"bids: {bids_dir}\n"
f"fmriprep: {fmriprep_dir}\n"
f"roi: {roi_dir}\n"
f"output: {output_dir}\n"
f"Debug: {debug}\n"
f"Regroup by domain: {regroup_by_domain}\n"
f"Save stat maps: {save_stat_maps}\n"
f"Save GLM report: {save_glm_report}\n"
f"Save weight maps: {save_weight_maps}\n"
f"Save confusion matrix: {save_confusion_matrix}\n"
f"Save final results as unique df: {save_final_results}\n"
)
from __future__ import annotations
import os
import re
import gc
import logging
import warnings
from datetime import datetime
from itertools import product
from pathlib import Path
from typing import Dict, Tuple, List, Any
import multiprocessing as _mp
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from nilearn.image import load_img
from nilearn.masking import apply_mask
from nilearn.glm.first_level import FirstLevelModel
from nilearn.maskers import NiftiMasker
from sklearn.svm import LinearSVC
from sklearn.preprocessing import StandardScaler
from sklearn.feature_selection import SelectPercentile, f_classif
from sklearn.pipeline import make_pipeline
from sklearn.metrics import accuracy_score, confusion_matrix
from sklearn.model_selection import LeavePGroupsOut, cross_val_score, cross_val_predict
from sklearn.base import clone
from joblib import Parallel, delayed, parallel_backend
warnings.filterwarnings("ignore", category=UserWarning)
warnings.filterwarnings("ignore", category=FutureWarning)
logging.getLogger("nilearn").setLevel(logging.ERROR)
# --------------- Parameters --------------
# GLM
hrf_model: str = "spm" # HRF function
high_pass: float = 0.008 # low cut filter
smoothing_fwhm: int = 4 # smoothing
# Decoder
n_leave_out: int = 2 #number of leave out
screening_percentile: int = 100 # purcentage of voxel kept
C: float = 1.0 #?
n_jobs_total: int = 20 # CPUs to use
# --------------- Parameters --------------
print("\n" + "-" * 40)
print(" ANALYSIS PARAMETERS")
print("-" * 40)
print("GLM parameters:")
print(f" • hrf_model : {hrf_model}")
print(f" • high_pass : {high_pass}")
print(f" • smoothing_fwhm : {smoothing_fwhm}")
print("Decoder parameters:")
print(f" • n_leave_out : {n_leave_out}")
print(f" • screening_pct : {screening_percentile}")
print(f" • Linear SVC C : {C}")
print(f" • Total CPUs : {n_jobs_total}")
print("-" * 40 + "\n")
def notify_save(description: str, path: os.PathLike | str) -> None: # noqa: N802
"""
prints a standardised message for each save
"""
print(f"Saved {description} to {path}")
def load_visual_events_by_run(events_dir: str, subject_id: str, task_name: str):
"""
load the events from presentation log for each run
"""
print(f"\nLoading visual events for sub-{subject_id}, keeping only img/objX events...")
events_dir = os.path.abspath(os.path.join(os.getcwd(), "..", data_location, "bids", f"sub-{subject_id}", "func"))
events: Dict[int, pd.DataFrame] = {}
pattern = re.compile(rf"sub-{subject_id}_task-{task_name}_run-(\d+)_events\.tsv")
obj_pattern = re.compile(r"(img/obj\d+)")
for filename in os.listdir(events_dir):
match = pattern.match(filename)
if match:
run_num = int(match.group(1))
filepath = os.path.join(events_dir, filename)
df = pd.read_csv(filepath, sep="\t")
if "trial_type" in df.columns:
if mri_trigger:
trigger_mask = df["trial_type"] == "MRI_trigger"
if trigger_mask.any():
trigger_onset = df.loc[trigger_mask, "onset"].iloc[0]
df["onset"] -= trigger_onset
df = df[df["trial_type"].str.startswith("img/obj", na=False)].copy()
df["trial_type"] = df["trial_type"].str.extract(obj_pattern)
if regroup_by_domain:
df["trial_type"] = df["trial_type"].replace(domain_mapping)
events[run_num] = df
conditions = events[1]["trial_type"].unique() if 1 in events else None
print(f"sub-{subject_id}: {len(events)} events files for {len(events)} runs loaded from {events_dir}.")
print(f"conditions: {conditions}")
return events, conditions
def load_bold_by_run(func_dir: str, subject_id: str, task_name: str):
"""
load bold data processed by fmriprep
"""
print(f"Loading functional data for sub-{subject_id} ...")
func: Dict[int, Any] = {}
pattern = re.compile(rf"sub-{subject_id}_task-{task_name}_run-(\d+)_echo-1_space-MNI152NLin2009cAsym_desc-preproc_bold\.nii\.gz")
for filename in os.listdir(func_dir):
match = pattern.match(filename)
if match:
run_num = int(match.group(1))
filepath = os.path.join(func_dir, filename)
func[run_num] = load_img(filepath)
print(f"sub-{subject_id}: {len(func)} functional files loaded from {func_dir}.")
return func
def find_nii_or_niigz(path_without_ext: str) -> str:
"""
returns existing .nii ou .nii.gz .
"""
nii_path = f"{path_without_ext}.nii"
niigz_path = f"{path_without_ext}.nii.gz"
if os.path.exists(nii_path):
return nii_path
if os.path.exists(niigz_path):
return niigz_path
raise FileNotFoundError(f"Neither '{nii_path}' nor '{niigz_path}' found.")
def compute_contrasts_by_run(
subject_id: str,
roi_name: str,
mask_img: str | os.PathLike,
func: Dict[int, Any],
events: Dict[int, pd.DataFrame],
runs: List[int],
):
z_maps, conditions_label, run_label = [], [], []
for run in runs:
try:
glm = FirstLevelModel(
t_r=t_r,
mask_img=mask_img,
hrf_model=hrf_model,
high_pass=high_pass,
smoothing_fwhm=smoothing_fwhm,
)
glm.fit(func[run], events=events[run])
for cond in events[run].trial_type.unique():
z_map = glm.compute_contrast(cond)
z_maps.append(z_map)
conditions_label.append(cond)
run_label.append(int(run))
safe_condition = cond.replace("/", "_")
# stat maps
if save_stat_maps:
outdir = Path(output_dir) / "stat_maps"
outdir.mkdir(parents=True, exist_ok=True)
filename = f"sub-{subject_id}_task-{task_name}_run-{run}_roi-{roi_name}_cond-{safe_condition}_zmap.nii.gz"
filepath = outdir / filename
z_map.to_filename(filepath)
notify_save("Z‑map", filepath)
# GLM report
if save_glm_report:
from nilearn.reporting import make_glm_report
report = make_glm_report(glm, contrasts=cond)
report_dir = Path(output_dir) / "glm_reports"
report_dir.mkdir(parents=True, exist_ok=True)
report_filename = (
f"sub-{subject_id}_task-{task_name}_run-{run}_roi-{roi_name}_cond-{safe_condition}_glmreport.html"
)
report_filepath = report_dir / report_filename
report.save_as_html(report_filepath)
notify_save("GLM report", report_filepath)
except Exception as e:
print(f"GLM failed sub-{subject_id} run-{run} ROI={roi_name}: {e}")
return z_maps, conditions_label, run_label
def fit_and_evaluate_decoder(
z_maps: List[Any],
mask_img: str | os.PathLike,
y: List[str],
groups: List[int],
screening_percentile: int = 100,
C: float = 1.0,
n_leave_out: int = 2,
n_jobs: int = 1,
):
# Vectorisation
z_maps = np.array(z_maps)
X = apply_mask(z_maps.reshape(-1, z_maps.shape[-1]), mask_img).astype("float32", copy=False)
# Pipeline
pipe = make_pipeline(
StandardScaler(),
SelectPercentile(f_classif, percentile=screening_percentile),
LinearSVC(C=C, dual=False),
)
# parallelised Cross‑validation
logo = LeavePGroupsOut(n_groups=n_leave_out)
with parallel_backend("loky", inner_max_num_threads=1):
scores = cross_val_score(pipe, X, y, groups=groups, cv=logo, n_jobs=n_jobs, verbose=0)
acc_mean, acc_std = scores.mean(), scores.std()
print(f" Accuracy L{n_leave_out}O: {acc_mean:.4f} ± {acc_std:.4f}")
pipe.fit(X, y)
# confusion matrix (no training)
y_true, y_pred = [], []
for train_idx, test_idx in logo.split(X, y, groups):
X_train, X_test = X[train_idx], X[test_idx]
y_train, y_test = np.array(y)[train_idx], np.array(y)[test_idx]
clf = clone(pipe)
clf.fit(X_train, y_train)
y_pred.extend(clf.predict(X_test))
y_true.extend(y_test)
cm = confusion_matrix(y_true, y_pred)
return acc_mean, 1 / len(np.unique(y)), scores.tolist(), cm, pipe
def process_subject_roi(
subject_id: str,
roi_name: str,
mask_path: str | os.PathLike,
unique_runs_by_sub: dict[str, list[int]]
):
try:
# runs specific to that sub
try:
runs_subset = unique_runs_by_sub[subject_id]
except KeyError:
raise ValueError(f"No runs of the expected type for sub-{subject_id}")
bids_func = os.path.join(bids_dir, f"sub-{subject_id}", "func")
preproc_func = os.path.join(fmriprep_dir, f"sub-{subject_id}", "func")
events, _ = load_visual_events_by_run(bids_func, subject_id, task_name)
func = load_bold_by_run(preproc_func, subject_id, task_name)
z_maps, labels, runs = compute_contrasts_by_run(
subject_id,
roi_name,
mask_path,
func,
events,
runs_subset
)
acc, chance, scores, cm, model = fit_and_evaluate_decoder(
z_maps,
mask_path,
labels,
runs,
screening_percentile=screening_percentile,
C=C,
n_leave_out=n_leave_out,
n_jobs=1, # here, keep 1 !
)
if save_stat_maps:
out_prefix = os.path.join(
output_dir,
"weight_maps",
f"sub-{subject_id}_roi-{roi_name}_task-{task_name}",
)
Path(out_prefix).parent.mkdir(parents=True, exist_ok=True)
save_decoder_weights(
model,
mask_path,
out_prefix,
class_labels=np.unique(labels).tolist(),
)
return (subject_id, roi_name), {
"accuracy": acc,
"chance_level": chance,
"accuracy_scores": scores,
"confusion_matrix": cm,
"conditions": np.unique(labels).tolist(),
"model": model,
}
except Exception as e:
print(f"Failed pipeline sub-{subject_id} ROI={roi_name}: {e}")
return (subject_id, roi_name), {}
def save_decoder_weights(
model,
mask_img: str | os.PathLike,
output_path_prefix: str,
class_labels: List[str] | None = None,
):
from nilearn.image import load_img
if isinstance(mask_img, (str, os.PathLike)):
mask_img = load_img(mask_img)
try:
svc = model.named_steps["linearsvc"]
selector = model.named_steps["selectpercentile"]
except Exception:
print("Could not extract 'linearsvc' or 'selectpercentile' from model — skipping weight maps.")
return
if not hasattr(svc, "coef_"):
print("Model SVC has no attribute 'coef_' — ensure it is trained and linear.")
return
coef = svc.coef_
if coef.ndim == 1:
coef = coef.reshape(1, -1)
n_classes = coef.shape[0]
support_mask = selector.get_support()
masker = NiftiMasker(mask_img=mask_img)
masker.fit()
for i in range(n_classes):
class_name = class_labels[i] if class_labels else f"class_{i}"
selected_weights = coef[i]
full_weights = np.zeros(support_mask.shape[0])
full_weights[support_mask] = selected_weights
# as nifiti
weights_img = masker.inverse_transform(full_weights)
nii_path = f"{output_path_prefix}_weights_{class_name}.nii.gz"
weights_img.to_filename(nii_path)
notify_save("weight map (NIfTI)", nii_path)
# as csv
mask_data = masker.mask_img_.get_fdata().astype(bool)
coords = np.column_stack(np.where(mask_data))
df = pd.DataFrame({"x": coords[:, 0], "y": coords[:, 1], "z": coords[:, 2], "weight": full_weights})
csv_path = f"{output_path_prefix}_weights_{class_name}.csv"
df.to_csv(csv_path, index=False)
notify_save("weights table (CSV)", csv_path)
if __name__ == "__main__":
os.makedirs(output_dir, exist_ok=True)
unique_runs = build_unique_runs_per_subject(
run_and_trial_types,
run_type=run_type,
n_runs_to_train_test=n_runs_to_train_test
)
roi_masks = {
roi: find_nii_or_niigz(os.path.join(roi_dir, roi))
for roi in roi_names
}
all_pairs = list(product(subject_list, roi_names))
with parallel_backend("loky", inner_max_num_threads=1):
results = Parallel(n_jobs=n_jobs_total, verbose=10)(
delayed(process_subject_roi)(
sub,
roi,
roi_masks[roi],
unique_runs
)
for sub, roi in all_pairs
)
decoding_results = dict(results)
group_suffix = "domains" if regroup_by_domain else "categories"
rows = []
for (sub, roi), res in decoding_results.items():
if not res:
continue
row = {
"subject": sub,
"roi": roi,
"accuracy_mean": res["accuracy"],
"chance_level": res["chance_level"],
}
for i, s in enumerate(res["accuracy_scores"]):
row[f"fold{i+1}"] = s
rows.append(row)
df_results = pd.DataFrame(rows)
runs_tag = f"{n_runs_to_train_test or 'all'}runs"
csv_name = (
f"decoding_accuracy_summary_{group_suffix}_{runs_tag}_L{n_leave_out}O_"
f"{datetime.today().strftime('%Y%m%d%H%M%S')}.csv"
)
csv_path = os.path.join(output_dir, csv_name)
df_results.to_csv(csv_path, index=False)
notify_save("decoding summary CSV", csv_path)
if save_confusion_matrix:
conf_dir = os.path.join(
output_dir,
f"conf_mat_{group_suffix}_{runs_tag}_L{n_leave_out}O"
)
os.makedirs(conf_dir, exist_ok=True)
for (sub, roi), res in decoding_results.items():
if not res:
continue
cm = res["confusion_matrix"]
conds = res["conditions"]
df_cm = pd.DataFrame(cm, index=conds, columns=conds)
cm_csv = os.path.join(conf_dir, f"sub-{sub}_roi-{roi}_cm.csv")
df_cm.to_csv(cm_csv)
notify_save("confusion matrix CSV", cm_csv)
plt.figure(figsize=(6, 5))
sns.heatmap(df_cm, annot=True, fmt="d", cmap="Blues")
plt.title(f"CM sub-{sub} ROI-{roi}")
plt.tight_layout()
cm_png = os.path.join(conf_dir, f"sub-{sub}_roi-{roi}_cm.png")
plt.savefig(cm_png)
plt.close()
notify_save("confusion matrix PNG", cm_png)
print("\nDONE !")
It is still very verbose with a lot of UserWaning and others in the print:
Exception ignored in: <function ResourceTracker.del at 0x7f2e5c75f9c0>
Traceback (most recent call last):
File “/home/touhara/anaconda3/envs/nilearn/lib/python3.13/multiprocessing/resource_tracker.py”, line 82, in del
File “/home/touhara/anaconda3/envs/nilearn/lib/python3.13/multiprocessing/resource_tracker.py”, line 91, in _stop
File “/home/touhara/anaconda3/envs/nilearn/lib/python3.13/multiprocessing/resource_tracker.py”, line 116, in _stop_locked
ChildProcessError: [Errno 10] No child processes
Also if I made any mistake in the script, please do not hesitate to notify me !
Matthieu