Summary of what happened:
Dear all,
I did a task in which participants rated ambiguous faces as happy or sad, and I added the response as a parametric modulator. In my events file, I have trial_type, events, duration, response. Some response lines were empty because participants did not answer to all stimuli. I created a column “modulation”. Since it was 0 or 1, I did this :
events1 = pd.read_csv(path_fMRI+sub+‘/func/’+sub+‘_task-priming_run-1_events.csv’)
events1[‘modulation’] = events1[‘response’] - events1[‘response’].mean()
events2 = pd.read_csv(path_fMRI+sub+‘/func/’+sub+‘_task-priming_run-2_events.csv’)
events2[‘modulation’] = events2[‘response’] - events2[‘response’].mean()
event_files = [events1, events2]
Then I’ve just made my design matrix like this:
design_matrices =
for idx, img in enumerate(fmri_imgs):
events = event_files[idx]
motion = movements[idx]
n_scans = img.shape[-1]
frame_times = np.arange(n_scans) * t_r
design_matrix = make_first_level_design_matrix(
frame_times,
events,
hrf_model=hrf_model,
drift_model=drift_model,
drift_order =drift_order,
add_regs=motion.values,
add_reg_names=motion.columns.tolist()
)
design_matrices.append(design_matrix)
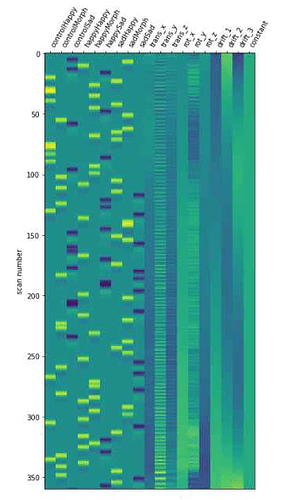
plot_design_matrix(design_matrices[1])
contrast_matrix = np.eye(design_matrix.shape[1])
conditions = {
column: contrast_matrix[i]
for i, column in enumerate(design_matrix.columns)
}
and know my modulation column has been taken into account because results make much more sense now that I added it and I’ve had this message: A ‘modulation’ column was found in the given events data and is used.
However, when I plot my design matrix, “modulation” does not appear! And I would like to extract the B of my modulator to see what drives how particpants respond to ambiguous faces as happy or sad, and have the psc in my ROIs!
Do you know if there is a way to extract it? Is there something wrong in the way I added the modulator?
Many thanks!!
Alexane