Does anyone know how to read the EXAR protocol files (export file format of protocols setup in Siemens scanner) from python to programmatically parse different steps and sequences inside them? Thanks.
I do not think these are documented or designed for public consumption. Perhaps someone else knows details. These are simply sqlite files, so reading the attributes is not hard, but interpreting them looks challenging. Here is the skeleton for a Python reader:
import sqlite3
con = sqlite3.connect('TERRA_20ch.exar1')
cur = con.cursor()
cur.execute("SELECT name FROM sqlite_master WHERE type='table';")
print(cur.fetchall())
cur.execute("SELECT * FROM Instance")
rows = cur.fetchall()
for row in rows:
print(row)
con.close()
Thanks Chris – I will give this a try! I was hoping someone here tried to do that already 
@Chris_Rorden Can you try that on your side:
#!/bin/env python3
#
# extract-exar1.py
#
# A small script to extract structures from SIEMENS exar1 file
# It will generate one JSON for each entry in the Content table.
# And sometimes an XML if json['Data'] attribute is found.
#
# Usage: python3 extract-exar1.py "filename.exar1"
#
# https://neurostars.org/t/parsing-exar-files/20237
import sqlite3
import sys
import zlib
import json
headers = [
'EDF V1: ContentType=syngo.MR.ExamDataFoundation.Data.EdfAddInConfigContent;',
'EDF V1: ContentType=syngo.MR.ExamDataFoundation.Data.EdfDecisionStepContent;',
'EDF V1: ContentType=syngo.MR.ExamDataFoundation.Data.EdfDirectoryContent;',
'EDF V1: ContentType=syngo.MR.ExamDataFoundation.Data.EdfInteractionStepContent;',
'EDF V1: ContentType=syngo.MR.ExamDataFoundation.Data.EdfJoinStepContent;',
'EDF V1: ContentType=syngo.MR.ExamDataFoundation.Data.EdfMeasurementStepContent;',
'EDF V1: ContentType=syngo.MR.ExamDataFoundation.Data.EdfPauseStepContent;',
'EDF V1: ContentType=syngo.MR.ExamDataFoundation.Data.EdfProgramContent;',
'EDF V1: ContentType=syngo.MR.ExamDataFoundation.Data.EdfProtocolContent;',
'EDF V1: ContentType=syngo.MR.ExamDataFoundation.Data.EdfSplitStepContent;',
'EDF V1: ContentType=syngo.MR.ExamDataFoundation.Data.EdfStringContent;',
'EDF V1: ContentType=syngo.MR.ExamDataFoundation.Data.EdfStructureContent;',
'EDF V1: ContentType=syngo.MR.ExamDataFoundation.Data.EdfWorkflowStepContent;'
]
filename = sys.argv[1]
table = 'Content'
con = sqlite3.connect(filename)
cur = con.cursor()
cur.execute(f"SELECT * FROM {table}")
rows = cur.fetchall()
for row in rows:
name = row[0] # unique hash ?
deflate_data = row[1]
assert deflate_data
assert row[2] == 'DS' # wotsit ?
unzipped = zlib.decompress(deflate_data, -zlib.MAX_WBITS)
str_data = unzipped.decode("utf-8")
# skip first line:
# 'EDF V1: ContentType=syngo.MR.ExamDataFoundation.Data.EdfAddInConfigContent;'
lines = str_data.splitlines(keepends=True)
header = lines[0].strip()
assert header in headers
json_str = ''.join(lines[1:])
# make sure this is JSON before writing it:
json_data = json.loads(json_str)
with open(name + '.json', 'w') as f1:
f1.write(json_str)
if 'Data' in json_data:
xml_data = json_data['Data']
# Always write with XML file extension, even for XProtocol
with open(name + '.xml', 'w') as f2:
f2.write(xml_data)
con.close()
Hi,
I have the same quenstion about te EXAR files. Would be nice if i can extract sequence information.
It would be nice if someone could help me with the implementation.
Of course I took action myself:
- I installed Python
- I installed VS Code
- I execute the code from @malaterre in VS code (i named it ‘Exar.py’).
But I probably need to make some adjustments/i did someting wrong.
When I run the script I get the following:
PS C:\Users\Arinda> & C:/Users/Arinda/AppData/Local/Programs/Python/Python312/python.exe "c:/Users/Arinda/Downloads/mri exar files/exar.py"
Traceback (most recent call last):
File "c:\Users\Arinda\Downloads\mri exar files\exar.py", line 34, in <module>
filename = sys.argv[1]
~~~~~~~~^^^
IndexError: list index out of range
PS C:\Users\Arinda>
What actions/adjustments do I need to do to make the code work?
I am very happy if someone can help me!
I have made some progress with chatgtp.
The code seems to be executed correctly, but I don’t see any json/xml files in the folder.
#!/bin/env python3
#
# extract-exar1.py
#
# A small script to extract structures from SIEMENS exar1 file
# It will generate one JSON for each entry in the Content table.
# And sometimes an XML if json['Data'] attribute is found.
#
# Usage: python3 extract-exar1.py "filename.exar1"
#
# https://neurostars.org/t/parsing-exar-files/20237
import sqlite3
import sys
import zlib
import json
headers = [
'EDF V1: ContentType=syngo.MR.ExamDataFoundation.Data.EdfAddInConfigContent;',
'EDF V1: ContentType=syngo.MR.ExamDataFoundation.Data.EdfDecisionStepContent;',
'EDF V1: ContentType=syngo.MR.ExamDataFoundation.Data.EdfDirectoryContent;',
'EDF V1: ContentType=syngo.MR.ExamDataFoundation.Data.EdfInteractionStepContent;',
'EDF V1: ContentType=syngo.MR.ExamDataFoundation.Data.EdfJoinStepContent;',
'EDF V1: ContentType=syngo.MR.ExamDataFoundation.Data.EdfMeasurementStepContent;',
'EDF V1: ContentType=syngo.MR.ExamDataFoundation.Data.EdfPauseStepContent;',
'EDF V1: ContentType=syngo.MR.ExamDataFoundation.Data.EdfProgramContent;',
'EDF V1: ContentType=syngo.MR.ExamDataFoundation.Data.EdfProtocolContent;',
'EDF V1: ContentType=syngo.MR.ExamDataFoundation.Data.EdfSplitStepContent;',
'EDF V1: ContentType=syngo.MR.ExamDataFoundation.Data.EdfStringContent;',
'EDF V1: ContentType=syngo.MR.ExamDataFoundation.Data.EdfStructureContent;',
'EDF V1: ContentType=syngo.MR.ExamDataFoundation.Data.EdfWorkflowStepContent;'
]
filename = r"C:\Users\Arinda\Downloads\mri exar files\Abdomen.exar1"
table = 'Content'
con = sqlite3.connect(filename)
cur = con.cursor()
cur.execute(f"SELECT * FROM {table}")
rows = cur.fetchall()
for row in rows:
name = row[0] # unique hash ?
deflate_data = row[1]
assert deflate_data
assert row[2] == 'DS' # wotsit ?
unzipped = zlib.decompress(deflate_data, -zlib.MAX_WBITS)
str_data = unzipped.decode("utf-8")
# skip first line:
# 'EDF V1: ContentType=syngo.MR.ExamDataFoundation.Data.EdfAddInConfigContent;'
lines = str_data.splitlines(keepends=True)
header = lines[0].strip()
assert header in headers
json_str = ''.join(lines[1:])
# make sure this is JSON before writing it:
json_data = json.loads(json_str)
with open(name + '.json', 'w', encoding='utf-8') as f1:
f1.write(json_str)
if 'Data' in json_data:
xml_data = json_data['Data']
# Always write with XML file extension, even for XProtocol
with open(name + '.xml', 'w') as f2:
f2.write(xml_data)
con.close()
I didn’t see any json or xml files. Maybe I’m too stupid for this. But I would love it if I could make it happen.
The terminal result are:
PS C:\Users\Arinda> & C:/Users/Arinda/AppData/Local/Programs/Python/Python312/python.exe "c:/Users/Arinda/Downloads/mri exar files/extract-exar1.py"```Hi @Wilbr , the way we solved it is via exporting the EXAR protocol files from the scanner as XML, and then reading them. Here is an implementation example: from mrQA
that depends on an easily customizable/extensible Class to handle Siemens protocol files from the protocol library:
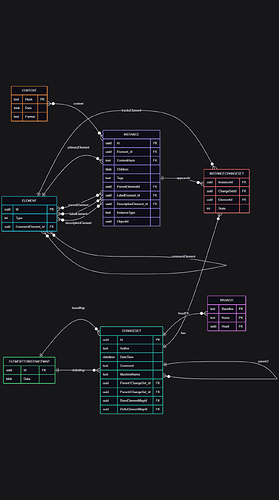
I found these were a little tricky to parse, however I’ve mapped out the database schema. Essentially every datatype is represented in the database as an “Instance” entry with corresponding content in the “Content” table. The “Element”, “InstanceChangeSet”, “ElementToInstanceMap”, and “Branch” tables don’t seem to be much use outside the scanner.
All of the data in each “Content” entry is stored as JSON (with a header describing the instance type). I found zlib.decompress can reliably decode the content BLOBs:
import zlib
def decode_content_min(b: bytes) -> bytes:
try:
return zlib.decompress(b, -15) # raw DEFLATE
except Exception:
return b # already plain or unexpected
One thing I did find is that the attributes that are neatly grouped into “Card” elements in the XML are not in the EXAR, and would require a fair bit of manual mapping to get them 1:1 with the content of the cards. You can find them in the very long “Data” string under the EdfProtocol instance’s JSON tied to each EdfMeasurementStep instance.