Summary of what happened:
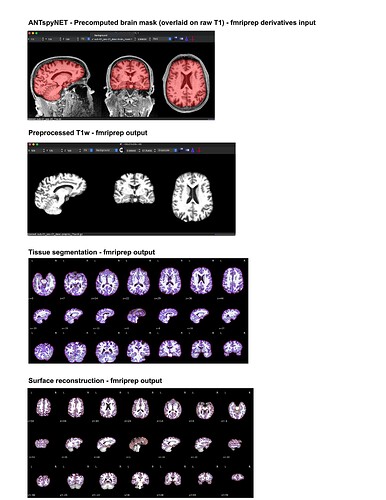
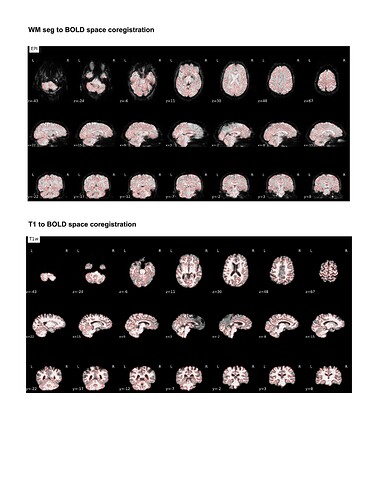
For my dataset I have found that ANTspyNET consistently produces a good/clean brain mask compared to fmriprep (antsBrainExtraction x Freesurfeer method). The default fmriprep method often outputs brain mask/segmentations that include the skull/non-brain tissues. As a result, I have been feeding my ANTspyNET mask as input via the --derivatives flag. While this works and the resulting outputs look good (see attached screenshots: decent tissue segmentations, good BOLD-T1 coreg etc), I noticed that the preprocessed anatomical T1 gets skull stripped in the process. Is this a sign of something going wrong under the hood?
Would greatly appreciate your thoughts, thank you!
Command used (and if a helper script was used, a link to the helper script or the command generated):
CMD="apptainer run --cleanenv \
-B /path_to_multiband_rsfMRI_data:/data \
${CONTAINER} \
/data/Outputs/BIDS \
/data/Outputs/fmriprep \
participant \
--participant-label 01 \
--session-label 01 \
--fs-license-file /data/Software/license.txt \
--force no-bbr \
--ignore slicetiming \
--output-spaces func anat MNI152NLin2009cAsym:res-native \
--skull-strip-t1w skip \
--derivatives fmriprep=/data/Outputs/fmriprep \
--use-syn-sdc \
-w /data/Outputs/fmriprep -vvv"
Version:
25.2.3
Environment (Docker, Singularity / Apptainer, custom installation):
Apptainer
Data formatted according to a validatable standard? Please provide the output of the validator:
251125-09:27:55,505 cli INFO:
Telemetry system to collect crashes and errors is enabled - thanks for your feedback!. Use option ``--notrack`` to opt out.
251125-09:27:59,596 cli DEBUG:
Initializing BIDS Layout
251125-09:28:45,171 nipype.workflow IMPORTANT:
Running fMRIPrep version 25.2.3
License NOTICE ##################################################
fMRIPrep 25.2.3
Copyright The NiPreps Developers.