I processed 3 separate, multiband fmri-bold runs through fMRIprep (task A: 2 runs, task B: 1 run, resting-state: 1 run).
PEPolar SDC was used. Task A shared one pair of spin-echo images, whereas task B and resting-state shared another pair.
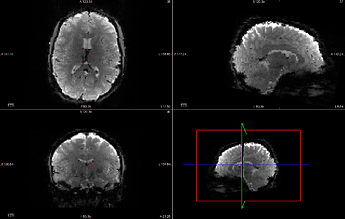
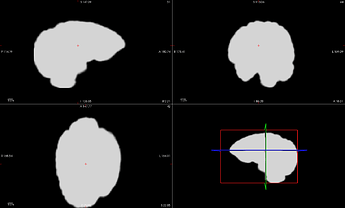
SDC applied to Task-A, run-1 clips and biases the image, ruining the extracted mask. This also ruins coregistration with the subject-space anatomical image.
original
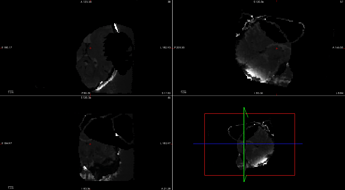
"corrected"
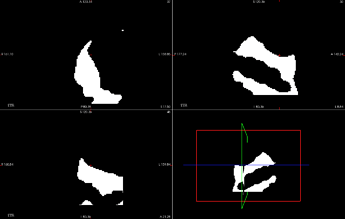
extracted mask
Task-A, run-1 and run-2 share the same spin-echo pair.
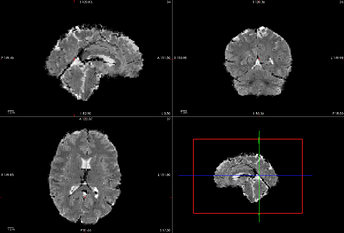
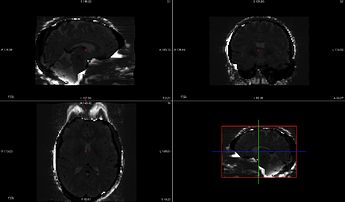
Below is an image of the estimated SDC, applied to Task-A, run-2.
Task-A run-2 Mask and original bold are unremarkable.
The SDC estimated for Task-B and resting-state are also unremarkable.
For Task-A, both runs, I confirmed good alignment between the sbref and original bold image. The "PhaseEncodingDirection" field was correctly applied. Might I ask if there are any other corrective procedures I can take?