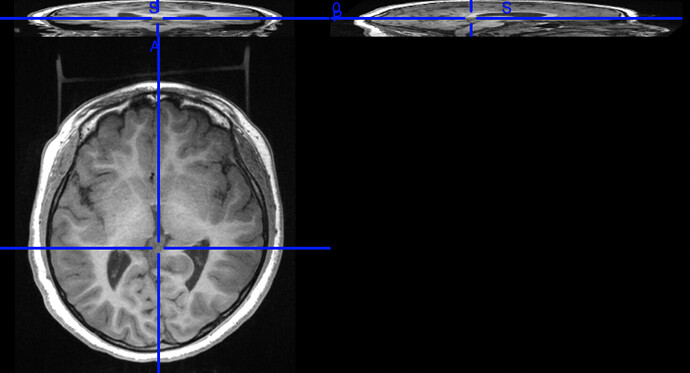
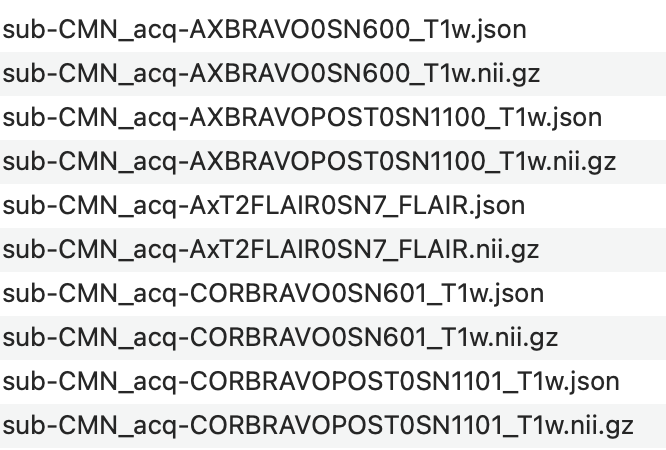
I have a nonconventional dataset, where I don’t have BOLD images but only multiple T1W images for each subject and one T2W image. Each T1W image “focuses” on one orientation. An example of how those images look like would be like the one shown below. Notice how the other two orientations are “squashed”. An example of one of the subject’s anat images is like this. The value of the acq keyword is composed of orientation (COR,AX,SAG), the technique used (i…e, BRAVO), and the serial number. For example, the first file’s name is sub-CMN_acq-AXBRAVO0SN600_T1w.nii, where AXBRAVO0SN600 stands for AX=Axial, BRAVO=technique used, 0=just to separate the technique from the serial number , and SN600=the serial number of the image. Some images have POST (like the 3rd file) which I am not sure what it refers to. But let us ignore this file for this moment.
I am interested in applying the anat pipeline of fmriprep but since I have multiple images, fmriprep will try (and I quote from docs):
For greater than two images, the cost can be a slowdown of an order of magnitude. Therefore, in the case of three or more images, fMRIPrep constructs templates aligned to the first image, unless passed the --longitudinal flag, which forces the estimation of an unbiased template.
I do not understand the behavior of the fmriprep in my case (because of different orientations). In perfect world, I hope that fmriprep will be able to join these 3 different orientations in a one template. Would it be able to do this? If not, how do you advice to run fmriprep based on this dataset.
I tried to run fmriprep on multiple subjects (with and without --longitudinal ) and I always get this error:
Saving crash info to /dir/BIDS-derivatives/sub-TJE/log/20220220-231522_741d0a9d-5b88-41a6-87a8-efe96de848b6/crash-20220220-231604-bakerh-t1w_ref_dimensions-b8fef8d5-006f-46a4-9666-0826a9a81684.txt
Traceback (most recent call last):
File "/opt/conda/lib/python3.8/site-packages/nipype/pipeline/plugins/multiproc.py", line 67, in run_node
result["result"] = node.run(updatehash=updatehash)
File "/opt/conda/lib/python3.8/site-packages/nipype/pipeline/engine/nodes.py", line 516, in run
result = self._run_interface(execute=True)
File "/opt/conda/lib/python3.8/site-packages/nipype/pipeline/engine/nodes.py", line 635, in _run_interface
return self._run_command(execute)
File "/opt/conda/lib/python3.8/site-packages/nipype/pipeline/engine/nodes.py", line 741, in _run_command
result = self._interface.run(cwd=outdir)
File "/opt/conda/lib/python3.8/site-packages/nipype/interfaces/base/core.py", line 428, in run
runtime = self._run_interface(runtime)
File "/opt/conda/lib/python3.8/site-packages/niworkflows/interfaces/images.py", line 447, in _run_interface
target_zooms = all_zooms[valid].min(axis=0)
File "/opt/conda/lib/python3.8/site-packages/numpy/core/_methods.py", line 43, in _amin
return umr_minimum(a, axis, None, out, keepdims, initial, where)
ValueError: zero-size array to reduction operation minimum which has no identity
220220-23:16:04,576 nipype.workflow INFO:
[Node] Setting-up "fmriprep_wf.single_subject_TJE_wf.anat_preproc_wf.fs_isrunning" in "/dir/fmriprep_wf/single_subject_TJE_wf/anat_preproc_wf/fs_isrunning".
220220-23:16:05,206 nipype.workflow INFO:
[Node] Running "fs_isrunning" ("nipype.interfaces.utility.wrappers.Function")
220220-23:16:05,250 nipype.workflow INFO:
[Node] Finished "fmriprep_wf.single_subject_TJE_wf.anat_preproc_wf.fs_isrunning".
220220-23:16:09,21 nipype.workflow INFO:
[Node] Finished "fmriprep_wf.single_subject_TJE_wf.anat_preproc_wf.brain_extraction_wf.lap_tmpl".
220220-23:16:10,472 nipype.workflow INFO:
[Node] Setting-up "fmriprep_wf.single_subject_TJE_wf.anat_preproc_wf.brain_extraction_wf.mrg_tmpl" in "/dir/fmriprep_wf/single_subject_TJE_wf/anat_preproc_wf/brain_extraction_wf/mrg_tmpl".
220220-23:16:10,623 nipype.workflow INFO:
[Node] Running "mrg_tmpl" ("nipype.interfaces.utility.base.Merge")
220220-23:16:10,664 nipype.workflow INFO:
[Node] Finished "fmriprep_wf.single_subject_TJE_wf.anat_preproc_wf.brain_extraction_wf.mrg_tmpl".
220220-23:16:14,410 nipype.workflow ERROR:
could not run node: fmriprep_wf.single_subject_TJE_wf.anat_preproc_wf.anat_template_wf.t1w_ref_dimensions
220220-23:16:14,421 nipype.workflow CRITICAL:
fMRIPrep failed: Workflow did not execute cleanly. Check log for details
220220-23:16:14,728 cli ERROR:
Preprocessing did not finish successfully. Errors occurred while processing data from participants: TJE (1). Check theHTML reports for details.
with different