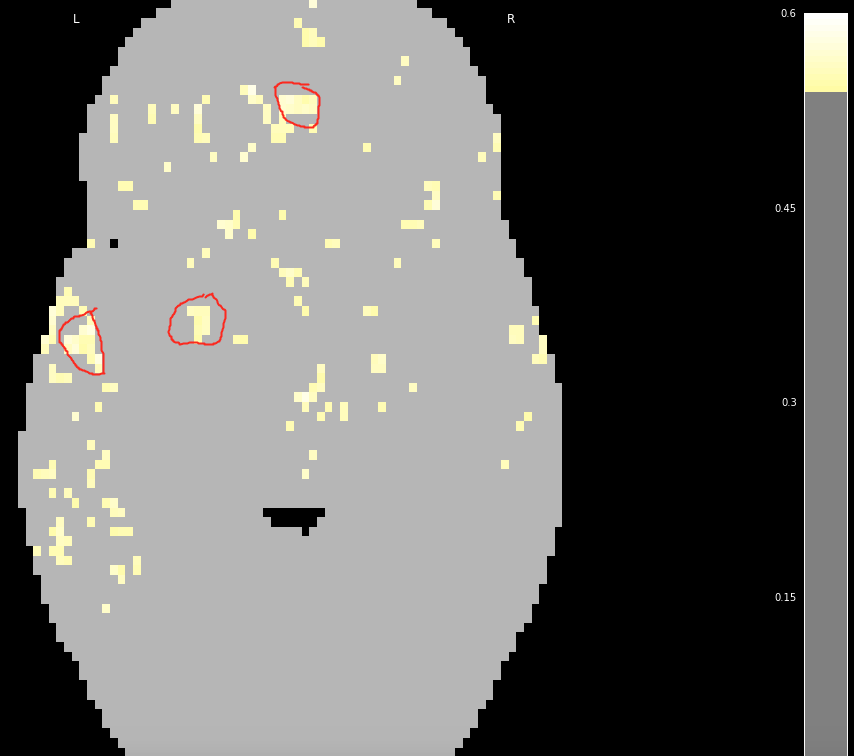
@slab15 below is a potential solution I came up with (assuming I understood what you want to do correctly…).
Note that there might be better ways to do this.
Let me know if this is of any help!
Nicolas
import numpy as np
from functools import reduce
from scipy.ndimage import label
from nibabel import Nifti1Image
from nilearn.image import binarize_img, get_data, new_img_like
# Creates a fake input image
# This would be equivalent to your
# thresholded stat map. I'm using
# a 2D image here because it's easier
# to see the clusters...
#
data = np.array([[0., 0., 0., 0., 0.],
[1., 0., 0., 1.1, 0.],
[0., 0., 0., 0., 2.],
[1., 0., 0., 1.2, 0.],
[2., 3., 0., 0., 0.]])
input_niimg = Nifti1Image(data, affine=np.eye(4))
def get_clusters_larger_than(niimg, min_size):
"""Returns a Nifti1Image where only clusters of
voxels of size larger than min_size were kept.
"""
data = get_data(binarize_img(niimg))
# structure defines the kind of connectivity
# that you wish to use
objs,n = label(data,
structure=np.ones(tuple([3] * len(data.shape)),
dtype=int))
cluster_sizes = np.bincount(objs.flatten())
labels_above_threshold = np.argwhere(cluster_sizes > min_size).flatten()[1:]
mask = reduce(np.logical_or, [objs == l for l in labels_above_threshold])
input_data = get_data(niimg)
input_data[~mask] = 0
return new_img_like(niimg, input_data)
output_niimg = get_clusters_larger_than(input_niimg, 2)
get_data(output_niimg)
array([[0. , 0. , 0. , 0. , 0. ],
[0. , 0. , 0. , 1.1, 0. ],
[0. , 0. , 0. , 0. , 2. ],
[1. , 0. , 0. , 1.2, 0. ],
[2. , 3. , 0. , 0. , 0. ]])