Summary of what happened:
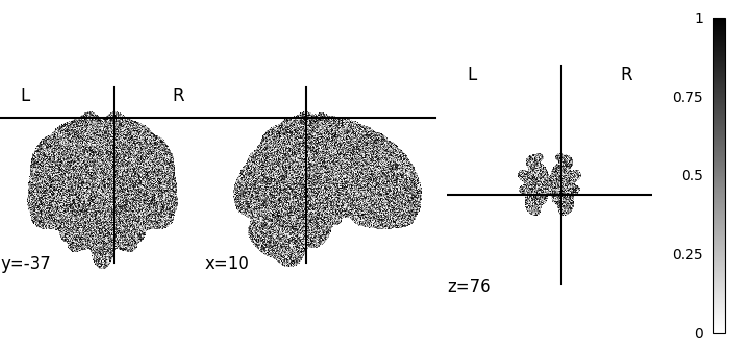
Hi, I’m new to nilearn. While trying to plot an image of the MNI152 template with all brain voxels set to white noise using nilearn.plotting.plot_img, I’m finding that the background values are always set to zero (and if I try setting the background voxel values to NaN, they get plotted as zero internally). If the colormap isn’t centered at zero, the background is then colored with something other than white, which is pretty much the only color I’d want for publication. Currently I’m hacking my way around this by separately tracking the indices of the background voxels from the masker and manually setting them to the value of the color I need for my particular colormap. Surely there’s a better way? I tried messing around with the bg_img option, but setting it to an image with the background voxels all set to the color I wanted didn’t do anything. From the docs I’m not sure how the bg_img interacts with the primary image.
Command used (and if a helper script was used, a link to the helper script or the command generated):
import numpy as np
from nilearn.input_data import NiftiMasker
import nibabel as nib
from nilearn import datasets, plotting
from nilearn.maskers import nifti_spheres_masker
from nibabel import Nifti1Image
template_image = datasets.load_mni152_template()
# mask_img = nib.load(mask_image_path)
# plotting.plot_img(mask_img, cut_coords=([-15, -10, -5, 0, 5]), display_mode='y', cmap='gray')
mask_img = datasets.load_mni152_brain_mask()
masker = NiftiMasker(mask_img=mask_img)
masker.fit()
# plotting.plot_roi(masker, template_image, alpha=0.5)
mask_data = mask_img.get_fdata()
affine = mask_img.affine
voxel_indices = np.array(np.where(mask_data > 0)).T
background_indices = np.where(mask_data == 0)
def voxel_to_atlas_coords(voxel_indices, affine):
"""Converts voxel indices to atlas coordinates."""
voxel_indices_homogeneous = np.c_[voxel_indices, np.ones(len(voxel_indices))]
world_coords = np.dot(affine, voxel_indices_homogeneous.T).T[:, :3]
return world_coords
voxel_coords = voxel_to_atlas_coords(voxel_indices, affine)
voxel_values = np.random.randn(voxel_coords.shape[0])
image_values = masker.inverse_transform(voxel_values)
assert len(voxel_indices) + len(background_indices[0]) == image_values.get_fdata().size
vals = image_values.get_fdata().copy()
# nilearn apparently pads with real values rather than NaNs... and also sets NaNs to zero if you try using them...
vals[background_indices] = vals.max()
plot_image = Nifti1Image(vals.copy(), affine=image_values.affine)
plotting.plot_img(plot_image,
colorbar=True,
cmap='gray',
)
Version:
Environment (Docker, Singularity / Apptainer, custom installation):
Data formatted according to a validatable standard? Please provide the output of the validator:
PASTE VALIDATOR OUTPUT HERE
Relevant log outputs (up to 20 lines):
PASTE LOG OUTPUT HERE