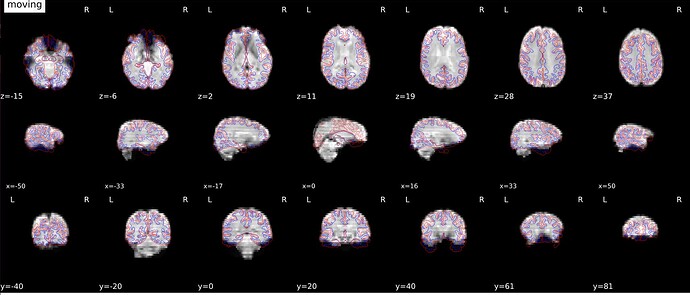
Dear all,
We used fmriprep-20.2.7 to preprocess our dataset, but we are facing issues with poor epi to T1w registration. Unfortunately, we do not have the field maps so we opted to use the option –use_syn_sdc in fmriprep. The registration results are quite poor between the EPI and T1 images, especially the frontal regions.
We tried different approaches below to solve this problem.
- As mentioned by prior post [Poor Registration EPI to T1w - #5 by Neer] (Poor Registration EPI to T1w - #5 by Neer) regarding the low of gray/white contrast, we tried to used FSL FLIRT command which is equivalent to --force-no-bbr in fmriprep to see if the registration works better. The registration results did not look any better.
> flirt -in /fmriprep/sub-CAMHLL032/ses-posttx/func/sub-CAMHLL032_ses-posttx_task-rest_boldref.nii.gz
-ref sub-CAMHLL032_skullstrip.nii.gz -dof 12 -cost bbr
-wmseg ./sub-CAMHLL032_label-WM_probseg.nii.gz -out ./sub-CAMH_taks-rest-postx_T1w.nii.gz
- We applied non-linear registration by using ANTSregistration SyN, which was suggested by this study (GitHub - NeuroanatomyAndConnectivity/epi_t1_nonlinear). The contrast intensity of T1w was inverted and matched to the EPI, and SyN deformation was restricted along the AP axis only. However, the results did not improve at all, they perhaps are even worse than previous outputs.
> `antsRegistration -g .1x1x.1
> --collapse-output-transforms 1 --dimensionality 3
> --initial-moving-transform [ /mnt/tigrlab/projects/ttan/ThirtyFourD/test_cores/test_output/sub-CAMHLL031/epi_t1_nonlinear/itk/epi2anat_affine.txt, 1 ]
> --initialize-transforms-per-stage 0
> --interpolation Linear --output [ transform, transform_Warped.nii.gz, transform_InverseWarped.nii.gz ]
> --transform SyN[ 0.1, 3.0, 0.0 ]
> --metric CC[ /mnt/tigrlab/projects/ttan/ThirtyFourD/test_cores/test_output/sub-CAMHLL031/epi_t1_nonlinear/maskepi/sub-CAMHLL031_ses-posttx_task-rest_desc-preproc_bold_mean_masked.nii.gz, /mnt/tigrlab/projects/ttan/ThirtyFourD/test_cores/test_output/sub-CAMHLL031/epi_t1_nonlinear/addinv/brain_out_masked_maths_maths.nii.gz, 1, 4, None ]
> --convergence [ 10x5, 1e-06, 10 ]
> --smoothing-sigmas 1.0x0.5vox --shrink-factors 2x1
> --use-estimate-learning-rate-once 1
> --use-histogram-matching 1 --winsorize-image-intensities [ 0.0, 1.0 ]
> --write-composite-transform 1`
- Currently, we are trying to implement the field-less map approach via sdcflows newest version https://www.nipreps.org/sdcflows/master/methods.html#fieldmap-less-approaches
We would really appreciate your expertise and comments on how to improve this poor EPI to T1w registration without the field maps.
Best,
T