Hi!
I am sometimes getting bad func/anatomical registration which results in low snr on parts of the brain where I wouldn’t expect to get low snr (like the lateral occipital lobe).
Here is the fMRIprep command I used.
singularity run --cleanenv -B /projects/:/projects/\
-B ${WORK_DIR}:/quest_scratch \
/projects/singularity_images/fmriprep-21.0.2.simg \
${BIDS_DIR} \
${BIDS_DIR}/${DERIVS_DIR} \
participant --participant-label ${subject} \
-w /quest_scratch --omp-nthreads 8 --nthreads 16 \
--fs-license-file /projects/singularity_images/freesurfer_license.txt \
--fs-subjects-dir ${BIDS_DIR}/${DERIVS_DIR}/sourcedata/freesurfer \
--output-spaces MNI152NLin6Asym:res-2 \
--ignore slicetiming --fd-spike-threshold 0.2 --me-output-echos --resource-monitor
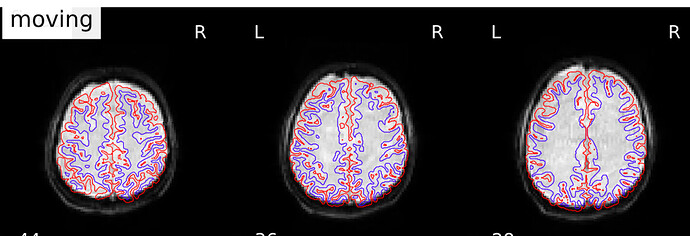
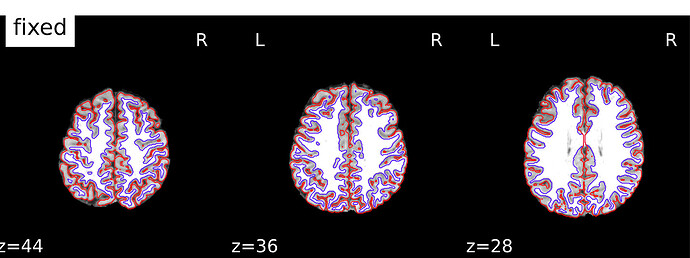
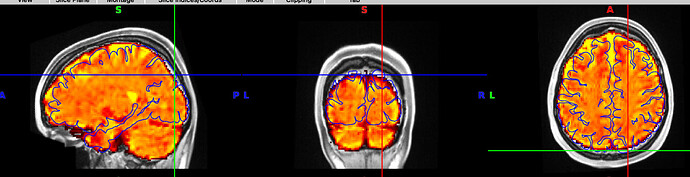
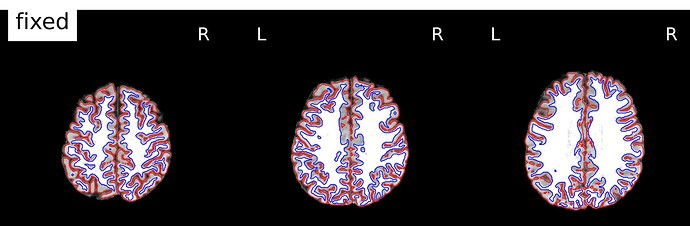
And here is an example where you can see the mismatch in the back of the brain, especially on the right hemisphere. The third image shows the boldref image on top of the T1 and the pial surface outline. no signal to be found.
Any idea why this is happening and how I can improve it?
1 Like
The gray/white contrast in your BOLD is extremely low, so boundary-based registration has little to work with. If you can find some other way to register it and update the header, you can use --bold2t1w-init header --force-no-bbr to tell fMRIPrep not to attempt any registration.
thank you @effigies
I do have single band reference files for each echo which all seem to have at least decent gray/white contrast (much better than what is shown in the registration screen shot i included in the last post).
Does fmriprep use them? From the documentation it looks like it should but then I saw some posts that sbrefs aren’t used for multiecho data?
I believe so but can’t check right now. Multi echo has definitely changed since 21.0. Can you try with 23.1?
1 Like
Hi @effigies
I tried it with 23.0.2 on a subset of sessions since we had that on our system already.
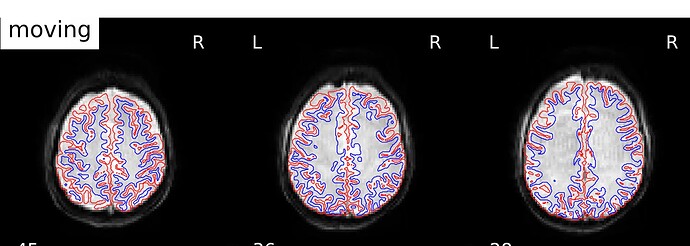
Registration looked a little better, but the contrast between gray/white matter in boldref images doesn’t look any better. What do you think?
How is it making these boldref images? The contrast is definitely much better in all the single band references I am passing in (at each echo)
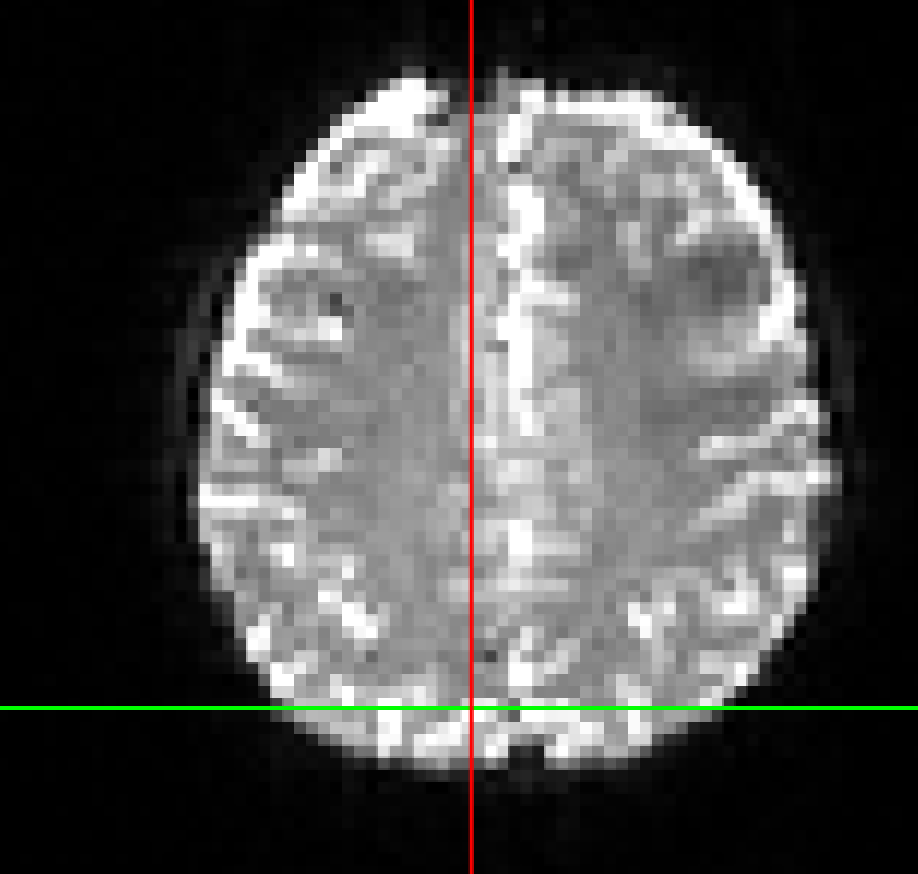
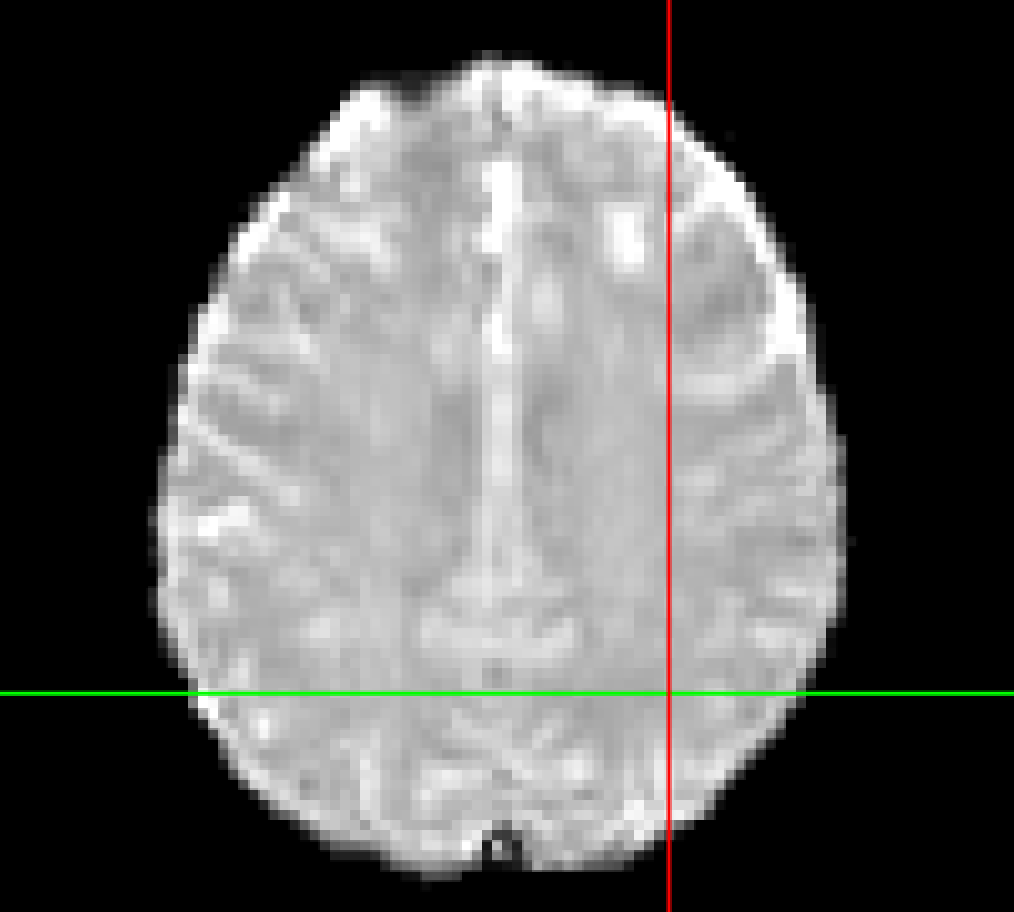
Here is an example of the SBref (echo 3) versus the boldref image for the same run
Hi @effigies ,
Just checking. Is your recommendation to register it elsewhere and then do fmriprep with --bold2t1w-init header --force-no-bbr?
Thanks for your patience. Yes, I think that is probably best. I looked at the code again, and we are using the sbrefs for the initial bold reference, which is used for head motion correction, but not for the final, which is used for coregistration. This needs fixing in an updated 23.2 series. That is unlikely to happen this month.
1 Like
jsein
July 12, 2023, 8:13pm
10
Interesting! On top of that, SBref images used as reference for HMC tend to bias the HMC parameters with the standard cost function used by mcflirt:
Hi, thank you for the fantastic work with FMRIPREP.
I just noticed something wrong in the output of FMRIPREP for a subject in a particular study: the motion parameters estimated by mcflirt are very strange, with a lot of zeros.
Here is the content of the .par file calculated by FMRIPREP v 20.2.1, launched through a singularity image on a cluster.
0 -0 0 0 0 0
0 -0 0 0 0 0.00447239
0 -0 0 0 0 0
0 -0 0 0 0 0.0191925
0 -0 0 0 0 0.0628555
0 -0 0 0 0 0.01720…
Wow, either I missed this or forgot about it. Have you found any issues in using mutualinfo for BOLD runs without sbrefs?
It does seem that we should probably prefer just to motion correct to the first volume and register the first volume to the reference, regardless of whether we use mcflirt or 3dvolreg.
jsein
July 12, 2023, 10:03pm
12
More details on this topic here:
opened 08:12PM - 23 Jan 21 UTC
bug
### What version of fMRIPrep are you using?
FMRIPREP v 20.2.1
### What kin… d of installation are you using? Containers (Singularity, Docker), or "bare-metal"?
Singularity installation (version 2.6.1-dist), running on a high performance computing cluster.
### What is the exact command-line you used?
```
singularity run -B /scratch/jsein/BIDS:/work,$HOME/.templateflow:/opt/templateflow --cleanenv /scratch/jsein/my_images/fmriprep-20.2.1.simg \
--fs-license-file /work/freesurfer/license.txt /work/$study /work/$study/derivatives \
participant --participant-label $list_sub \
-w /work/temp_data_${study}\
--mem-mb 50000 --omp-nthreads 10 --nthreads 12 --ignore slicetiming --fd-spike-threshold 0.5 \
--output-spaces MNI152NLin2009cAsym MNI152NLin6Asym --return-all-components --cifti-output 91k
```
### Have you checked that your inputs are BIDS valid?
yes the input passes the bids-validator
### Did fMRIPrep generate the visual report for this particular subject? If yes, could you share it?
<!-- we can download links from Dropbox, Box, Google Drive, etc. You can send them privately to nipreps@gmail.com.
Reports do not contain data usable with personal identification or other research purposes -->
Yes the visual report was generated, no error reported, it is under the dropbox link at the end of this post.
### Can you find some traces of the error reported in the visual report (at the bottom) or in *crashfiles*?
no error reported
### Are you reusing previously computed results (e.g., FreeSurfer, Anatomical derivatives, work directory of previous run)?
No
### fMRIPrep log
If you have access to the output logged by fMRIPrep, please make sure to attach it as a text file to this issue.
We observed this strange output while building the GLM with nuisance regressors and noting that the columns with zeros where correlated with each other.
output: HTML reports, output and confounds_timeseries.tsv files under the following link:
https://www.dropbox.com/sh/3e455qbh8anqwl3/AAACn2-tnmRBhXx44M_vO0qEa?dl=0
Here is for example the content of the .par file output of mcflirt run by fmriprep: the zeros are very strange and it is a very different result from what I obtain if I use SPM for realigning the same run (but realignment is to the first scan instead to the sbref as it is set with fmriprep) No zeros are found with SPM.
EDIT: I just check with my local FSL installation and I found that mcflirt is giving the same output as fmriprep! Thus it is not seem to be a problem with fmriprep, rather a strange behaviour of mcflirt!
This subject had very low movements.
Thank you for sharing your thoughts!
```
0 -0 0 0 0 0
0 -0 0 0 0 0
0 -0 0 0 0 0
0 -0 0 0 0 0
0 -0 0 0 0 0.0298349
0 -0 0 0 0 0
0 -0 0 0 0 0
0 -0 0 0 0 0.0264596
0 -0 0 0 0 0.0125837
0 -0 0 0 0 0.00908859
0 -0 0 0 0 0.0499751
0 -0 0 0 0 0.0285046
0 -0 0 0 0 0.0394745
0 -0 0 0 0 0.032549
0 -0 0 0 0 0.0142394
0 -0 0 0 0 0.0188951
0 -0 0 0 0 0.0527396
0 -0 0 0 0 0.0118122
0 -0 0 0 0 0.0324686
-0.00020877 -0 0 0 0.00806986 0.0617947
-0.00071251 -0 0 0 0.0156193 0.0508352
0 -0 0 0 0 0.0259631
0 -0 0 0 0 0.0267916
0 -0 0 0 0 0.0423937
0 -0 0 0 0 0.0269359
0 -0 0 0 0 0.0480931
0 -0 0 0 0 0.0533892
-0.00079703 -0 0 0 0.0137821 0.0708697
-0.000779751 -0 0 0 0.011465 0.0502716
-0.00123065 -0 0 0 0.010698 0.109888
-0.00137164 -0 0 -7.29355e-09 0.0228613 0.0814645
-0.000797153 -0 0 -1.14295e-08 0.00402441 0.0433854
-0.00012957 -0 0 0 -0.000157725 0.0451129
-9.30172e-05 -0 0 0 -0.000177668 0.0566566
-0.000718193 -0 0 2.98428e-09 0.0150825 0.070767
-0.000450858 -0 0 9.52406e-09 0.00859951 0.0501525
-0.000182812 -0 0 0 0.00272729 0.0540445
-0.000355515 -0 0 0 0.00660644 0.056919
-0.00093645 -0 0 8.85655e-09 0.022218 0.0617246
-0.00130357 -0 0 0 0.0175132 0.0757637
-0.000789502 -0 0 0 0.0188825 0.0440254
-0.000264678 -0 0 0 0.00222485 0.0428122
-0.000721302 -0 0 0 0.0046294 0.0479397
-0.000891793 -0 0 8.90645e-09 0.021491 0.063626
-0.000721302 -0 0 0 0.0129002 0.0448448
-0.000112873 -0 0 0 -0.000384434 0.0460485
-0.000721302 -0 0 0 0.0136793 0.0609756
-0.000918056 -0 0 -4.79318e-09 0.0214994 0.0758632
-0.000987211 -0 0 1.42662e-08 0.0215346 0.0572181
-1.62193e-05 -0 0 1.0704e-09 -0.0050121 0.0533982
-0.000114733 -0 0 1.0704e-09 -0.000784969 0.0627668
-0.000738586 -0 0 1.0704e-09 0.0136945 0.0650518
-0.000980801 -0 0 9.64596e-09 0.0215313 0.0748123
-0.000738586 -0 0 1.0704e-09 0.00835188 0.0467987
-0.000738586 -0 0 1.0704e-09 0.00858199 0.0601027
-0.000738586 -0 0 1.0704e-09 0.00791957 0.027476
-0.000915425 -0 0 -3.87668e-09 0.0215165 0.0781238
-0.000632387 -0 0 1.07851e-08 0.010477 0.0423524
-0.00106174 -0 0 1.0704e-09 0.0250179 0.0660661
-0.000252066 -0 0 1.0704e-09 0.00448506 0.015156
-0.000550322 -0 0 1.0704e-09 0.00689665 0.0323721
-0.000738587 -0 0 1.0704e-09 0.0154435 0.0451829
-0.000738587 -0 0 1.0704e-09 0.0157021 0.0451829
-0.000683615 -0 0 8.63665e-09 0.00997629 0.0415626
-0.000738587 -0 0 1.0704e-09 0.0144991 0.0451829
-0.000738587 -0 0 1.0704e-09 0.0214398 0.062077
-0.000846131 -0 0 5.43473e-09 0.0214808 0.0616804
-0.000855447 -0 0 1.0704e-09 0.0142566 0.0805584
-0.000772026 -0 0 -1.47155e-09 0.021433 0.0404133
-0.000692231 -0 0 1.21844e-09 0.00489802 0.0434587
-0.000692231 -0 0 1.21844e-09 0.0135194 0.0451152
-0.000947193 -0 0 1.04547e-08 0.0275542 0.0769438
-0.000776082 -0 0 7.4812e-09 0.014442 0.0472611
-0.000674085 -0 0 2.25856e-09 0.00508311 0.0388478
-0.000674085 -0 0 2.25856e-09 0.0121347 0.0496988
-0.000787968 -0 0 -1.61307e-10 0.0214144 0.0552333
-0.000126402 -0 0 2.25856e-09 -0.00279745 0.0390678
-0.000793037 -0 0 7.2322e-09 0.021416 0.0833968
-0.000849837 -0 0 -8.19674e-09 0.0214422 0.0486355
-0.000752162 -0 0 2.25856e-09 0.0111124 0.0452097
-0.000752162 -0 0 2.99291e-09 0.0130734 0.0452097
-0.000962554 -0 0 2.99291e-09 0.0214808 0.0777942
-0.000142289 -0 0 2.99291e-09 0.0018386 0.0489621
-0.000752162 -0 0 2.99291e-09 0.0131606 0.0601034
-0.00089234 -0 0 -6.56433e-09 0.0214494 0.0828999
-0.000888647 -0 0 -6.16855e-09 0.0157029 0.0803104
-0.000844328 -0 0 -1.78585e-09 0.011968 0.0887311
-0.00169953 -0 0 -7.47431e-09 0.0334674 0.158842
-0.00087393 -0 0 5.37133e-09 0.0146743 0.0623504
0.000143044 -0 0 2.99291e-09 -0.00734077 0.0754622
-0.000691344 -0 0 -9.25198e-09 0.00984836 0.0752114
-0.000752162 -0 0 2.99291e-09 0.016751 0.084167
-0.000752163 -0 0 2.99291e-09 0.0157413 0.083586
-0.000752163 -0 0 2.99291e-09 0.00653493 0.0807244
-8.70316e-05 -0 0 2.99291e-09 0.00169775 0.0917994
-9.26849e-05 -0 0 2.99291e-09 0.00127143 0.0895586
-0.000752163 -0 0 2.99291e-09 0.017261 0.0870561
-0.00125371 -0 0 -1.00326e-08 0.0288015 0.14875
-0.000752163 -0 0 2.99291e-09 0.0146633 0.0639313
-0.000118441 -0 0 2.99291e-09 0.00127299 0.0817119
-0.000752163 -0 0 2.99291e-09 0.0129707 0.0823899
-0.000752163 -0 0 2.99291e-09 0.0213944 0.106449
-0.000752163 -0 0 2.99291e-09 0.0113494 0.0763368
-0.00065891 -0 0 -8.25317e-09 0.0111491 0.0822857
-0.000671937 -0 0 2.99291e-09 0.0112994 0.0823012
-0.000752163 -0 0 2.99291e-09 0.0128503 0.0925025
-0.000752163 -0 0 2.99291e-09 0.0155407 0.0901279
-0.000752163 -0 0 2.99291e-09 0.0168779 0.113742
-0.00089253 -0 0 7.76998e-09 0.0214438 0.0883661
-0.000679152 -0 0 1.24104e-08 0.00631072 0.0723094
-0.000657444 -0 0 -6.72726e-09 0.0106853 0.0822833
-0.000689144 -0 0 -3.30677e-09 0.0124616 0.0823191
-0.000925563 -0 0 7.72727e-09 0.0214587 0.108435
-0.000698344 -0 0 8.31619e-11 0.0157116 0.072331
-0.00068413 -0 0 -8.76388e-09 0.0117933 0.0863004
-0.000752164 -0 0 2.99291e-09 0.0161003 0.0883442
-0.000920858 -0 0 -5.52268e-09 0.0214595 0.113374
-0.000752164 -0 0 2.99291e-09 0.0147892 0.0723899
-0.000917793 -0 0 2.99291e-09 0.0214578 0.10781
-0.000689913 -0 0 2.29434e-10 0.0149052 0.0711225
-0.000566403 -0 0 9.45319e-09 0.00925237 0.0821727
-0.000752164 -0 0 2.99291e-09 0.0213944 0.0962025
-0.000752164 -0 0 2.99291e-09 0.0213944 0.0823899
-0.000638266 -0 0 1.16246e-08 0.0110292 0.0722563
-0.00066447 -0 0 2.99291e-09 0.0136406 0.0880102
-0.000752164 -0 0 2.99291e-09 0.0213944 0.0947306
-0.000651978 -0 0 -2.81807e-09 0.011058 0.0689206
-0.000667694 -0 0 1.15758e-08 0.0139397 0.0877071
-0.00154329 -0 0 2.99291e-09 0.0401142 0.136862
-0.00129329 -0 0 9.52764e-09 0.0260016 0.0759717
-0.000107472 -0 0 2.99291e-09 0.000271841 0.0816564
-2.47086e-05 -0 0 2.99291e-09 0.000230031 0.0877171
-5.01933e-05 -0 0 2.99291e-09 0.000241494 0.105102
-5.01933e-05 -0 0 2.99291e-09 0.000238228 0.0815856
-0.000111185 -0 0 2.99291e-09 0.000261973 0.0979274
-0.000300193 -0 0 2.99291e-09 0.00312824 0.113329
-0.000147117 -0 0 2.99291e-09 0.00027752 0.0988603
-5.01934e-05 -0 0 2.99291e-09 0.000244518 0.101356
-0.000482272 -0 0 2.99291e-09 0.0106453 0.139763
-5.01935e-05 -0 0 2.99291e-09 0.000247003 0.0815848
-0.000139604 -0 0 2.99291e-09 0.000282857 0.0957424
-0.000491681 -0 0 -1.30105e-08 0.0116485 0.119675
-5.01935e-05 -0 0 2.99291e-09 0.000251651 0.0815925
-7.66508e-05 -0 0 2.99291e-09 0.000259712 0.0977094
-5.01936e-05 -0 0 2.99291e-09 0.000249538 0.112486
-0.000558213 -0 0 2.99291e-09 0.0109131 0.112771
-5.01937e-05 -0 0 2.99291e-09 0.000246456 0.0977375
-0.000300194 -0 0 2.99291e-09 0.00514414 0.122534
-0.00134459 -0 0 1.54441e-08 0.0328532 0.141568
-0.000300194 -0 0 2.99291e-09 0.00815844 0.0883218
-0.000183202 -0 0 2.99291e-09 0.000293394 0.102353
-4.32239e-05 -0 0 2.99291e-09 0.000238674 0.107906
-0.00114743 -0 0 2.99291e-09 0.0290794 0.127425
-0.000897429 -0 0 1.00482e-08 0.0173483 0.0952883
-0.000889111 -0 0 1.04315e-08 0.0214945 0.105775
-0.00100408 -0 0 9.48222e-09 0.019849 0.11848
-0.00127335 -0 0 5.11865e-09 0.0311654 0.130845
-0.000296748 -0 0 2.08935e-09 0.00617523 0.0873417
-0.000969976 -0 0 1.85995e-09 0.0236238 0.109871
-0.00105306 -0 0 -8.34191e-10 0.0290198 0.101678
-0.000337284 -0 0 2.94065e-09 0.00976135 0.0935801
-0.000912522 -0 0 2.94065e-09 0.0217164 0.106609
-0.00100806 -0 0 1.09586e-08 0.0290003 0.0980294
-0.000235892 -0 0 2.94065e-09 0.00416129 0.0680161
-0.0002995 -0 0 -4.24463e-09 0.00763374 0.0718959
-0.000378003 -0 0 2.94065e-09 0.00766427 0.09784
-0.000476403 -0 0 1.142e-08 0.0105637 0.107273
-0.000264467 -0 0 1.70832e-08 0.00462611 0.0818563
-0.000378003 -0 0 2.94065e-09 0.00766427 0.107528
-0.00127529 -0 0 2.94065e-09 0.0279142 0.115822
-0.000394514 -0 0 2.94065e-09 0.0054216 0.0635719
-0.000338767 -0 0 2.94065e-09 0.00535792 0.0620108
-0.000310327 -0 0 2.94065e-09 0.00973431 0.0701747
-0.000510893 -0 0 -1.20349e-08 0.0121424 0.0721408
-0.000306212 -0 0 2.94065e-09 0.00654379 0.0608874
-0.000307715 -0 0 2.94065e-09 0.00973395 0.0745652
-0.000760893 -0 0 2.94065e-09 0.0166699 0.0886181
-0.000402248 -0 0 2.94065e-09 0.00976776 0.0587203
-0.00037274 -0 0 2.94065e-09 0.0097618 0.0704316
-0.00133981 -0 0 -7.1319e-09 0.0341621 0.118327
-0.00114327 -0 0 8.29037e-09 0.0243787 0.061372
-0.000397666 -0 0 1.7511e-09 0.00560246 0.048239
-0.000467144 -0 0 1.7511e-09 0.0135842 0.0720942
-0.000703884 -0 0 1.7511e-09 0.0136784 0.0925856
-0.000567805 -0 0 1.42559e-08 0.0136245 0.068412
-0.000456984 -0 0 1.7511e-09 0.0082714 0.082086
-0.00049742 -0 0 1.7511e-09 0.013601 0.0875239
-0.000579684 -0 0 6.21471e-09 0.0136322 0.0943948
-0.000886482 -0 0 -1.90109e-09 0.0178705 0.132457
-0.000392981 -0 0 1.7511e-09 0.00837947 0.0465562
-0.000453884 -0 0 1.7511e-09 0.00354411 0.0682447
-0.000455754 -0 0 1.7511e-09 0.00778869 0.076941
-0.00079791 -0 0 -2.25216e-09 0.0160182 0.119483
-0.000703884 -0 0 1.7511e-09 0.0136784 0.0860687
-0.000515806 -0 0 8.14077e-09 0.00505693 0.0721474
-0.000703884 -0 0 1.7511e-09 0.0136784 0.109038
-0.000973579 -0 0 1.7511e-09 0.0191724 0.0930738
-0.000703884 -0 0 1.7511e-09 0.00652437 0.102228
-0.00137622 -0 0 -3.02707e-09 0.0290181 0.121431
-0.000922695 -0 0 1.7511e-09 0.0164082 0.0597458
-0.000535682 -0 0 -1.52291e-08 0.00822229 0.0487303
-0.000416535 -0 0 1.7511e-09 0.0056854 0.0698911
-0.000675995 -0 0 1.52494e-08 0.0160388 0.101727
-0.000494376 -0 0 1.7511e-09 0.0135606 0.0598621
-0.000427305 -0 0 1.7511e-09 0.00713759 0.0711866
-0.00042642 -0 0 1.7511e-09 0.00950132 0.0675095
-0.000545192 -0 0 -1.2961e-08 0.0135841 0.0911589
-0.00113445 -0 0 4.96017e-09 0.0287818 0.0692903
-0.000248316 -0 0 1.7511e-09 0.0026122 0.0552848
-0.000296377 -0 0 1.7511e-09 0.00952237 0.0707775
-0.000984914 -0 0 -9.48523e-09 0.0220887 0.0928407
-0.00108519 -0 0 1.7511e-09 0.0242357 0.0728167
-0.000428385 -0 0 1.7511e-09 0.00972028 0.0664081
-0.000440844 -0 0 1.7511e-09 0.00669708 0.0700118
-0.000577341 -0 0 1.43718e-08 0.0147366 0.0949765
-0.000714186 -0 0 -8.42742e-09 0.0147911 0.0766545
-0.000567001 -0 0 1.7511e-09 0.00955145 0.0650231
-0.000952613 -0 0 1.7511e-09 0.0198898 0.107762
-0.000711122 -0 0 1.25251e-08 0.0147255 0.0502363
-0.000141859 -0 0 2.83079e-09 0.00133122 0.00681592
-0.000254764 -0 0 2.83079e-09 0.00438108 0.0245639
-0.000573799 -0 0 2.83079e-09 0.0144983 0.0500758
-0.000396892 -0 0 2.83079e-09 0.00894718 0.036291
-0.000450751 -0 0 1.14495e-08 0.0105671 0.0509599
-0.000477195 -0 0 -4.98444e-09 0.0144614 0.0619916
-0.000765275 -0 0 -2.22546e-09 0.0165784 0.104925
-0.000498349 -0 0 6.246e-09 0.00804579 0.0667286
-0.000573799 -0 0 2.83079e-09 0.00861331 0.107492
-0.000479679 -0 0 -4.89206e-09 0.00855949 0.0620016
-0.000323799 -0 0 2.83079e-09 0.00669057 0.0618326
-0.000505582 -0 0 1.70776e-08 0.0144763 0.0964717
-0.000483779 -0 0 -3.67766e-09 0.00838124 0.0620033
-0.000383789 -0 0 2.83079e-09 0.00331371 0.0780429
-0.000474073 -0 0 -5.8192e-10 0.00939916 0.0801106
-0.000573799 -0 0 2.83079e-09 0.0144983 0.0963335
-0.000475001 -0 0 -4.62251e-10 0.00791038 0.0719908
-0.000573799 -0 0 2.83079e-09 0.0144983 0.106045
-0.000484343 -0 0 -3.61993e-10 0.0144627 0.0481019
-0.000323799 -0 0 2.83079e-09 0.0056614 0.0513834
-0.00038702 -0 0 1.12477e-08 0.00775919 0.0662933
-0.000468504 -0 0 2.83079e-09 0.0144591 0.0814031
-0.000381009 -0 0 2.83079e-09 0.00618318 0.0502864
-0.000323799 -0 0 2.83079e-09 0.00439705 0.0645692
-0.000484721 -0 0 2.83079e-09 0.0144647 0.0776531
-0.000483708 -0 0 6.24865e-09 0.0144612 0.0556088
-0.000323799 -0 0 2.83079e-09 0.0102559 0.0534578
-0.000508544 -0 0 -8.55307e-09 0.0144758 0.0826264
-0.000475607 -0 0 2.83079e-09 0.0144603 0.055945
-0.00048232 -0 0 -8.61192e-10 0.0144635 0.0668524
-0.000573799 -0 0 2.83079e-09 0.0144983 0.055117
-0.000323799 -0 0 2.83079e-09 0.00819916 0.0483753
-0.000323799 -0 0 2.83079e-09 0.00439067 0.0618151
-0.000573799 -0 0 2.83079e-09 0.0144983 0.085265
-0.000477475 -0 0 2.83079e-09 0.0144557 0.045981
-0.000390461 -0 0 -6.07088e-09 0.00444682 0.0553409
-0.000477257 -0 0 6.35485e-09 0.0113832 0.0619901
-0.000750476 -0 0 1.25869e-08 0.0197093 0.107349
-0.000481706 -0 0 -3.52031e-10 0.0144592 0.0509564
-0.000323799 -0 0 2.83079e-09 0.0103062 0.055793
-0.00046289 -0 0 2.83079e-09 0.014456 0.0756933
-0.000495254 -0 0 8.29451e-09 0.0144655 0.0462053
-0.000373825 -0 0 2.83079e-09 0.0108125 0.051343
-0.000573799 -0 0 2.83079e-09 0.0144983 0.0721011
-0.000481413 -0 0 5.48884e-09 0.0144615 0.0403123
-0.000378721 -0 0 2.83079e-09 0.00595095 0.0527988
-0.000573799 -0 0 2.83079e-09 0.0144983 0.102255
-0.000400382 -0 0 -4.43367e-09 0.0069007 0.0287226
6.30042e-05 -0 0 2.83079e-09 -0.0026651 0.0452635
-0.000435495 -0 0 2.83079e-09 0.0097738 0.0547686
-0.000469106 -0 0 2.83079e-09 0.00982248 0.0560717
0.000125741 -0 0 2.83079e-09 -0.00412512 0.0484292
-0.000462934 -0 0 2.83079e-09 0.00751252 0.0650845
-0.000667804 -0 0 -5.24346e-09 0.0145207 0.0721987
-0.000424832 -0 0 3.74969e-09 0.00839867 0.0542887
-0.00050323 -0 0 3.74969e-09 0.0144516 0.083276
-0.00118624 -0 0 3.74969e-09 0.0289049 0.10322
-0.000578415 -0 0 3.74969e-09 0.0157659 0.0492656
-0.000412126 -0 0 3.74969e-09 0.00332497 0.054508
-0.000404368 -0 0 3.74969e-09 0.0123336 0.0578388
-0.00051215 -0 0 9.10393e-09 0.0123738 0.0910823
-0.000404368 -0 0 3.74969e-09 0.00882121 0.0543322
-0.000404368 -0 0 3.74969e-09 0.0123336 0.0539334
-0.000404368 -0 0 3.74969e-09 0.00871471 0.0561
-0.000404368 -0 0 3.74969e-09 0.00873877 0.0555488
-0.000507047 -0 0 -5.76854e-09 0.0123691 0.0897949
-0.000768395 -0 0 1.24991e-08 0.0158443 0.0859467
-0.000392008 -0 0 2.32531e-09 0.00623554 0.0343081
-0.000392008 -0 0 2.32531e-09 0.0122711 0.0456019
-0.000392008 -0 0 2.32531e-09 0.0122711 0.0526101
-0.000392008 -0 0 2.32531e-09 0.00931002 0.0517353
-0.000392008 -0 0 2.32531e-09 0.00584972 0.0423322
-0.000392008 -0 0 2.32531e-09 0.0122711 0.0527413
-0.000392008 -0 0 2.32531e-09 0.0122711 0.0619527
-0.000537575 -0 0 -2.55388e-09 0.012324 0.0721128
-0.000457688 -0 0 -3.89321e-10 0.0122959 0.0840514
-0.000474748 -0 0 -6.42473e-10 0.0123049 0.0461615
-0.000392008 -0 0 2.32531e-09 0.00542904 0.0464291
-0.000392008 -0 0 2.32531e-09 0.0122711 0.0504269
-0.000392008 -0 0 2.32531e-09 0.0122711 0.0719527
-0.000392008 -0 0 2.32531e-09 0.0122711 0.0507589
-0.000392008 -0 0 2.32531e-09 0.00858723 0.0541215
-0.000392008 -0 0 2.32531e-09 0.008138 0.0656587
-0.000551162 -0 0 2.32531e-09 0.0123354 0.0721295
-0.000392008 -0 0 2.32531e-09 0.00491105 0.0497302
-0.000482046 -0 0 -6.23686e-10 0.0123049 0.0720546
-0.0006911 -0 0 2.32531e-09 0.0194784 0.0669943
-0.000360711 -0 0 2.32531e-09 0.00555064 0.0250721
-0.000347486 -0 0 2.32531e-09 0.00468455 0.0332049
-0.000388303 -0 0 2.32531e-09 0.0122163 0.0419559
0.000231651 -0 0 2.32531e-09 -0.00607035 0.0548363
-0.000304605 -0 0 2.32531e-09 0.0079979 0.0580075
-0.000170488 -0 0 2.32531e-09 0.0029905 0.0312546
0.000271837 -0 0 -1.80617e-09 -0.00468443 0.0380728
0.000268027 -0 0 1.17999e-08 -0.00618964 0.0445663
0.000261084 -0 0 1.35479e-08 -0.00598816 0.0452548
0.000181061 -0 0 2.32531e-09 -0.00862791 0.0828628
0.00036988 -0 0 2.32531e-09 -0.0119828 0.0501614
0.00027085 -0 0 1.25974e-08 -0.00556844 0.0679252
0.000181061 -0 0 2.32531e-09 -0.0060663 0.0913342
0.000276382 -0 0 -2.61899e-09 -0.0119468 0.0343135
0.000274122 -0 0 2.32531e-09 -0.00934721 0.0487565
0.000322834 -0 0 1.18787e-08 -0.0061962 0.0650543
0.000181061 -0 0 2.32531e-09 -0.00699194 0.0828496
0.000181061 -0 0 2.32531e-09 -0.00716909 0.0417544
0.000339541 -0 0 -5.74701e-09 -0.00958281 0.0612099
-0.000146911 -0 0 2.32531e-09 0.00494863 0.105116
0.000181061 -0 0 2.32531e-09 -0.00365363 0.0208984
0.000288511 -0 0 2.32531e-09 -0.0119517 0.0223716
0.00027053 -0 0 6.85316e-09 -0.00629294 0.0500681
-0.000236648 -0 0 2.32531e-09 0.00532296 0.0606858
0.000181061 -0 0 2.32531e-09 -0.00860397 0.0279844
0.000244789 -0 0 -2.63327e-09 -0.00729157 0.0500972
2.38631e-06 -0 0 2.32531e-09 0.00281907 0.0794415
0.000181061 -0 0 2.32531e-09 -0.00350589 0.0501742
0.000181061 -0 0 2.32531e-09 -0.00955198 0.0625052
-2.97965e-06 -0 0 2.32531e-09 0.00210593 0.0949698
0.000181061 -0 0 2.32531e-09 -0.00830979 0.043568
0.000298899 -0 0 -5.45857e-09 -0.0119505 0.0534515
0.000181061 -0 0 2.32531e-09 -0.00190411 0.0874287
0.000181061 -0 0 2.32531e-09 -0.00374029 0.0634447
0.000317519 -0 0 2.32531e-09 -0.0119585 0.0529064
0.000181061 -0 0 2.32531e-09 -0.00312477 0.0772102
7.06293e-05 -0 0 2.32531e-09 0.00037782 0.0733328
0.000289854 -0 0 7.07189e-09 -0.00840966 0.0473801
0.000331191 -0 0 -5.55191e-09 -0.0119579 0.0599957
-0.000241041 -0 0 2.32531e-09 0.0050688 0.0806819
0.000181061 -0 0 2.32531e-09 -0.00190411 0.0167831
0.000181061 -0 0 2.32531e-09 -0.00844981 0.0195735
0.000340188 -0 0 2.32531e-09 -0.0119715 0.0361546
0.000181061 -0 0 2.32531e-09 -0.00597024 0.031775
0.000181061 -0 0 2.32531e-09 -0.00214056 0.0480441
0.000181061 -0 0 2.32531e-09 -0.00407231 0.015303
0.000181061 -0 0 2.32531e-09 -0.00520885 0.0195735
0 -0 0 0 0 0.0343214
```
Here is a plot (made with FSLeyes) of the translation along x, calculated with mcflirt (.par file, 4th column, ref= sbref) and with spm (rp file, 1st column, ref=1st scan): Quite different!

Another example: same run, looking at the ortation around y, calculated with mcflirt (.par file, 2nd column, ref= sbref) and with spm (rp file, 5th column, ref=1st scan):

Discussion with FSL developpers here:https://www.jiscmail.ac.uk/cgi-bin/wa-jisc.exe?A2=ind2101&L=FSL&D=0&X=5CD9DAA0225F506E17&Y=julien.sein%40univ-amu.fr&P=190219
Discussion on the HCPPipelines list here:
opened 11:18PM - 24 Jan 21 UTC
Hi, I am not exactely a HCPPipelines user but rather a FMRIPREP user but I notic… ed an issue with a methodology that I was derived from the one used by the HCP pipeline, so I thought I might as well post my question here:
For a subject with low movements (FDmean < 0.1mm for TR=1.1s) I noticed that the .par file output from mcflirt used by FMRIPREP was filled with many zeros, which is strange:
```
0 -0 0 0 0 0
0 -0 0 0 0 0
0 -0 0 0 0 0
0 -0 0 0 0 0
0 -0 0 0 0 0.0298349
0 -0 0 0 0 0
0 -0 0 0 0 0
0 -0 0 0 0 0.0264596
0 -0 0 0 0 0.0125837
0 -0 0 0 0 0.00908859
0 -0 0 0 0 0.0499751
0 -0 0 0 0 0.0285046
0 -0 0 0 0 0.0394745
0 -0 0 0 0 0.032549
0 -0 0 0 0 0.0142394
0 -0 0 0 0 0.0188951
0 -0 0 0 0 0.0527396
0 -0 0 0 0 0.0118122
0 -0 0 0 0 0.0324686
-0.00020877 -0 0 0 0.00806986 0.0617947
-0.00071251 -0 0 0 0.0156193 0.0508352
0 -0 0 0 0 0.0259631
0 -0 0 0 0 0.0267916
0 -0 0 0 0 0.0423937
0 -0 0 0 0 0.0269359
0 -0 0 0 0 0.0480931
0 -0 0 0 0 0.0533892
-0.00079703 -0 0 0 0.0137821 0.0708697
-0.000779751 -0 0 0 0.011465 0.0502716
-0.00123065 -0 0 0 0.010698 0.109888
-0.00137164 -0 0 -7.29355e-09 0.0228613 0.0814645
-0.000797153 -0 0 -1.14295e-08 0.00402441 0.0433854
-0.00012957 -0 0 0 -0.000157725 0.0451129
-9.30172e-05 -0 0 0 -0.000177668 0.0566566
-0.000718193 -0 0 2.98428e-09 0.0150825 0.070767
-0.000450858 -0 0 9.52406e-09 0.00859951 0.0501525
-0.000182812 -0 0 0 0.00272729 0.0540445
-0.000355515 -0 0 0 0.00660644 0.056919
-0.00093645 -0 0 8.85655e-09 0.022218 0.0617246
-0.00130357 -0 0 0 0.0175132 0.0757637
-0.000789502 -0 0 0 0.0188825 0.0440254
-0.000264678 -0 0 0 0.00222485 0.0428122
-0.000721302 -0 0 0 0.0046294 0.0479397
-0.000891793 -0 0 8.90645e-09 0.021491 0.063626
-0.000721302 -0 0 0 0.0129002 0.0448448
-0.000112873 -0 0 0 -0.000384434 0.0460485
-0.000721302 -0 0 0 0.0136793 0.0609756
-0.000918056 -0 0 -4.79318e-09 0.0214994 0.0758632
-0.000987211 -0 0 1.42662e-08 0.0215346 0.0572181
-1.62193e-05 -0 0 1.07039e-09 -0.0050121 0.0533982
-0.000114733 -0 0 1.07039e-09 -0.000784969 0.0627668
-0.000738586 -0 0 1.07039e-09 0.0136945 0.0650518
-0.000980801 -0 0 9.64594e-09 0.0215313 0.0748123
-0.000738586 -0 0 1.07039e-09 0.00835188 0.0467987
-0.000738586 -0 0 1.07039e-09 0.00858199 0.0601027
-0.000738586 -0 0 1.07039e-09 0.00791957 0.027476
-0.000915425 -0 0 -3.87669e-09 0.0215165 0.0781238
-0.000632387 -0 0 1.07851e-08 0.010477 0.0423524
-0.00106174 -0 0 1.07039e-09 0.0250179 0.0660661
-0.000252066 -0 0 1.07039e-09 0.00448506 0.015156
-0.000550322 -0 0 1.07039e-09 0.00689665 0.0323721
-0.000738587 -0 0 1.07039e-09 0.0154435 0.0451829
-0.000738587 -0 0 1.07039e-09 0.0157021 0.0451829
-0.000683615 -0 0 8.63663e-09 0.00997629 0.0415626
-0.000738587 -0 0 1.07039e-09 0.0144991 0.0451829
-0.000738587 -0 0 1.07039e-09 0.0214398 0.062077
-0.000846131 -0 0 5.43471e-09 0.0214808 0.0616804
-0.000855447 -0 0 1.07039e-09 0.0142566 0.0805584
-0.000772026 -0 0 -1.47156e-09 0.021433 0.0404133
-0.000692231 -0 0 1.21842e-09 0.00489802 0.0434587
-0.000692231 -0 0 1.21842e-09 0.0135194 0.0451152
-0.000947193 -0 0 1.04546e-08 0.0275542 0.0769438
-0.000776082 -0 0 7.48119e-09 0.014442 0.0472611
-0.000674085 -0 0 2.25855e-09 0.00508311 0.0388478
-0.000674085 -0 0 2.25855e-09 0.0121347 0.0496988
-0.000787968 -0 0 -1.61322e-10 0.0214144 0.0552333
-0.000126402 -0 0 2.25855e-09 -0.00279745 0.0390678
-0.000793037 -0 0 7.23219e-09 0.021416 0.0833968
-0.000849837 -0 0 -8.19674e-09 0.0214422 0.0486355
-0.000752162 -0 0 2.25855e-09 0.0111124 0.0452097
-0.000752162 -0 0 2.99289e-09 0.0130734 0.0452097
-0.000962554 -0 0 2.99289e-09 0.0214808 0.0777942
-0.000142289 -0 0 2.99289e-09 0.0018386 0.0489621
-0.000752162 -0 0 2.99289e-09 0.0131606 0.0601034
-0.00089234 -0 0 -6.56435e-09 0.0214494 0.0828999
-0.000888647 -0 0 -6.16856e-09 0.0157029 0.0803104
-0.000844328 -0 0 -1.78586e-09 0.011968 0.0887311
-0.00169953 -0 0 -7.47433e-09 0.0334674 0.158842
-0.00087393 -0 0 5.37132e-09 0.0146743 0.0623504
0.000143044 -0 0 2.99289e-09 -0.00734077 0.0754622
-0.000691344 -0 0 -9.25199e-09 0.00984836 0.0752114
-0.000752162 -0 0 2.99289e-09 0.016751 0.084167
-0.000752163 -0 0 2.99289e-09 0.0157413 0.083586
-0.000752163 -0 0 2.99289e-09 0.00653493 0.0807244
-8.70316e-05 -0 0 2.99289e-09 0.00169775 0.0917994
-9.26849e-05 -0 0 2.99289e-09 0.00127143 0.0895586
-0.000752163 -0 0 2.99289e-09 0.017261 0.0870561
-0.00125371 -0 0 -1.00326e-08 0.0288015 0.14875
-0.000752163 -0 0 2.99289e-09 0.0146633 0.0639313
-0.000118441 -0 0 2.99289e-09 0.00127299 0.0817119
-0.000752163 -0 0 2.99289e-09 0.0129707 0.0823899
-0.000752163 -0 0 2.99289e-09 0.0213944 0.106449
-0.000752163 -0 0 2.99289e-09 0.0113494 0.0763368
-0.00065891 -0 0 -8.25318e-09 0.0111491 0.0822857
-0.000671937 -0 0 2.99289e-09 0.0112994 0.0823012
-0.000752163 -0 0 2.99289e-09 0.0128503 0.0925025
-0.000752163 -0 0 2.99289e-09 0.0155407 0.0901279
-0.000752163 -0 0 2.99289e-09 0.0168779 0.113742
-0.00089253 -0 0 7.76997e-09 0.0214438 0.0883661
-0.000679152 -0 0 1.24104e-08 0.00631072 0.0723094
-0.000657444 -0 0 -6.72726e-09 0.0106853 0.0822833
-0.000689144 -0 0 -3.30678e-09 0.0124616 0.0823191
-0.000925563 -0 0 7.72725e-09 0.0214587 0.108435
-0.000698344 -0 0 8.31477e-11 0.0157116 0.072331
-0.00068413 -0 0 -8.76389e-09 0.0117933 0.0863004
-0.000752164 -0 0 2.99289e-09 0.0161003 0.0883442
-0.000920858 -0 0 -5.52269e-09 0.0214595 0.113374
-0.000752164 -0 0 2.99289e-09 0.0147892 0.0723899
-0.000917793 -0 0 2.99289e-09 0.0214578 0.10781
-0.000689913 -0 0 2.29434e-10 0.0149052 0.0711225
-0.000566403 -0 0 9.45317e-09 0.00925237 0.0821727
-0.000752164 -0 0 2.99289e-09 0.0213944 0.0962025
-0.000752164 -0 0 2.99289e-09 0.0213944 0.0823899
-0.000638266 -0 0 1.16245e-08 0.0110292 0.0722563
-0.00066447 -0 0 2.99289e-09 0.0136406 0.0880102
-0.000752164 -0 0 2.99289e-09 0.0213944 0.0947306
-0.000651978 -0 0 -2.81808e-09 0.011058 0.0689206
-0.000667694 -0 0 1.15758e-08 0.0139397 0.0877071
-0.00154329 -0 0 2.99289e-09 0.0401142 0.136862
-0.00129329 -0 0 9.52762e-09 0.0260016 0.0759717
-0.000107472 -0 0 2.99289e-09 0.000271841 0.0816564
-2.47086e-05 -0 0 2.99289e-09 0.000230031 0.0877171
-5.01933e-05 -0 0 2.99289e-09 0.000241494 0.105102
-5.01933e-05 -0 0 2.99289e-09 0.000238228 0.0815856
-0.000111185 -0 0 2.99289e-09 0.000261973 0.0979274
-0.000300193 -0 0 2.99289e-09 0.00312824 0.113329
-0.000147117 -0 0 2.99289e-09 0.00027752 0.0988603
-5.01934e-05 -0 0 2.99289e-09 0.000244518 0.101356
-0.000482272 -0 0 2.99289e-09 0.0106453 0.139763
-5.01935e-05 -0 0 2.99289e-09 0.000247003 0.0815848
-0.000139604 -0 0 2.99289e-09 0.000282857 0.0957424
-0.000491681 -0 0 -1.30105e-08 0.0116485 0.119675
-5.01935e-05 -0 0 2.99289e-09 0.000251651 0.0815925
-7.66508e-05 -0 0 2.99289e-09 0.000259712 0.0977094
-5.01936e-05 -0 0 2.99289e-09 0.000249538 0.112486
-0.000558213 -0 0 2.99289e-09 0.0109131 0.112771
-5.01937e-05 -0 0 2.99289e-09 0.000246456 0.0977375
-0.000300194 -0 0 2.99289e-09 0.00514414 0.122534
-0.00134459 -0 0 1.54441e-08 0.0328532 0.141568
-0.000300194 -0 0 2.99289e-09 0.00815844 0.0883218
-0.000183202 -0 0 2.99289e-09 0.000293394 0.102353
-4.32239e-05 -0 0 2.99289e-09 0.000238674 0.107906
-0.00114743 -0 0 2.99289e-09 0.0290794 0.127425
-0.000897429 -0 0 1.00482e-08 0.0173483 0.0952883
-0.000889111 -0 0 1.04315e-08 0.0214945 0.105775
-0.00100408 -0 0 9.48221e-09 0.019849 0.11848
-0.00127335 -0 0 5.11864e-09 0.0311654 0.130845
-0.000296748 -0 0 2.08934e-09 0.00617523 0.0873417
-0.000969976 -0 0 1.85993e-09 0.0236238 0.109871
-0.00105306 -0 0 -8.34206e-10 0.0290198 0.101678
-0.000337284 -0 0 2.94064e-09 0.00976135 0.0935801
-0.000912522 -0 0 2.94064e-09 0.0217164 0.106609
-0.00100806 -0 0 1.09586e-08 0.0290003 0.0980294
-0.000235892 -0 0 2.94064e-09 0.00416129 0.0680161
-0.0002995 -0 0 -4.24464e-09 0.00763374 0.0718959
-0.000378003 -0 0 2.94064e-09 0.00766427 0.09784
-0.000476403 -0 0 1.14199e-08 0.0105637 0.107273
-0.000264467 -0 0 1.70831e-08 0.00462611 0.0818563
-0.000378003 -0 0 2.94064e-09 0.00766427 0.107528
-0.00127529 -0 0 2.94064e-09 0.0279142 0.115822
-0.000394514 -0 0 2.94064e-09 0.0054216 0.0635719
-0.000338767 -0 0 2.94064e-09 0.00535792 0.0620108
-0.000310327 -0 0 2.94064e-09 0.00973431 0.0701747
-0.000510893 -0 0 -1.20349e-08 0.0121424 0.0721408
-0.000306212 -0 0 2.94064e-09 0.00654379 0.0608874
-0.000307715 -0 0 2.94064e-09 0.00973395 0.0745652
-0.000760893 -0 0 2.94064e-09 0.0166699 0.0886181
-0.000402248 -0 0 2.94064e-09 0.00976776 0.0587203
-0.00037274 -0 0 2.94064e-09 0.0097618 0.0704316
-0.00133981 -0 0 -7.13192e-09 0.0341621 0.118327
-0.00114327 -0 0 8.29036e-09 0.0243787 0.061372
-0.000397666 -0 0 1.75109e-09 0.00560246 0.048239
-0.000467144 -0 0 1.75109e-09 0.0135842 0.0720942
-0.000703884 -0 0 1.75109e-09 0.0136784 0.0925856
-0.000567805 -0 0 1.42559e-08 0.0136245 0.068412
-0.000456984 -0 0 1.75109e-09 0.0082714 0.082086
-0.00049742 -0 0 1.75109e-09 0.013601 0.0875239
-0.000579684 -0 0 6.21469e-09 0.0136322 0.0943948
-0.000886482 -0 0 -1.9011e-09 0.0178705 0.132457
-0.000392981 -0 0 1.75109e-09 0.00837947 0.0465562
-0.000453884 -0 0 1.75109e-09 0.00354411 0.0682447
-0.000455754 -0 0 1.75109e-09 0.00778869 0.076941
-0.00079791 -0 0 -2.25218e-09 0.0160182 0.119483
-0.000703884 -0 0 1.75109e-09 0.0136784 0.0860687
-0.000515806 -0 0 8.14076e-09 0.00505693 0.0721474
-0.000703884 -0 0 1.75109e-09 0.0136784 0.109038
-0.000973579 -0 0 1.75109e-09 0.0191724 0.0930738
-0.000703884 -0 0 1.75109e-09 0.00652437 0.102228
-0.00137622 -0 0 -3.02708e-09 0.0290181 0.121431
-0.000922695 -0 0 1.75109e-09 0.0164082 0.0597458
-0.000535682 -0 0 -1.52292e-08 0.00822229 0.0487303
-0.000416535 -0 0 1.75109e-09 0.0056854 0.0698911
-0.000675995 -0 0 1.52494e-08 0.0160388 0.101727
-0.000494376 -0 0 1.75109e-09 0.0135606 0.0598621
-0.000427305 -0 0 1.75109e-09 0.00713759 0.0711866
-0.00042642 -0 0 1.75109e-09 0.00950132 0.0675095
-0.000545192 -0 0 -1.2961e-08 0.0135841 0.0911589
-0.00113445 -0 0 4.96016e-09 0.0287818 0.0692903
-0.000248316 -0 0 1.75109e-09 0.0026122 0.0552848
-0.000296377 -0 0 1.75109e-09 0.00952237 0.0707775
-0.000984914 -0 0 -9.48525e-09 0.0220887 0.0928407
-0.00108519 -0 0 1.75109e-09 0.0242357 0.0728167
-0.000428385 -0 0 1.75109e-09 0.00972028 0.0664081
-0.000440844 -0 0 1.75109e-09 0.00669708 0.0700118
-0.000577341 -0 0 1.43718e-08 0.0147366 0.0949765
-0.000714186 -0 0 -8.42742e-09 0.0147911 0.0766545
-0.000567001 -0 0 1.75109e-09 0.00955145 0.0650231
-0.000952613 -0 0 1.75109e-09 0.0198898 0.107762
-0.000711122 -0 0 1.25251e-08 0.0147255 0.0502363
-0.000141859 -0 0 2.83077e-09 0.00133122 0.00681592
-0.000254764 -0 0 2.83077e-09 0.00438108 0.0245639
-0.000573799 -0 0 2.83077e-09 0.0144983 0.0500758
-0.000396892 -0 0 2.83077e-09 0.00894718 0.036291
-0.000450751 -0 0 1.14495e-08 0.0105671 0.0509599
-0.000477195 -0 0 -4.98446e-09 0.0144614 0.0619916
-0.000765275 -0 0 -2.22548e-09 0.0165784 0.104925
-0.000498349 -0 0 6.24598e-09 0.00804579 0.0667286
-0.000573799 -0 0 2.83077e-09 0.00861331 0.107492
-0.000479679 -0 0 -4.89207e-09 0.00855949 0.0620016
-0.000323799 -0 0 2.83077e-09 0.00669057 0.0618326
-0.000505582 -0 0 1.70775e-08 0.0144763 0.0964717
-0.000483779 -0 0 -3.67767e-09 0.00838124 0.0620033
-0.000383789 -0 0 2.83077e-09 0.00331371 0.0780429
-0.000474073 -0 0 -5.81935e-10 0.00939916 0.0801106
-0.000573799 -0 0 2.83077e-09 0.0144983 0.0963335
-0.000475001 -0 0 -4.62251e-10 0.00791038 0.0719908
-0.000573799 -0 0 2.83077e-09 0.0144983 0.106045
-0.000484343 -0 0 -3.62007e-10 0.0144627 0.0481019
-0.000323799 -0 0 2.83077e-09 0.0056614 0.0513834
-0.00038702 -0 0 1.12477e-08 0.00775919 0.0662933
-0.000468504 -0 0 2.83077e-09 0.0144591 0.0814031
-0.000381009 -0 0 2.83077e-09 0.00618318 0.0502864
-0.000323799 -0 0 2.83077e-09 0.00439705 0.0645692
-0.000484721 -0 0 2.83077e-09 0.0144647 0.0776531
-0.000483708 -0 0 6.24864e-09 0.0144612 0.0556088
-0.000323799 -0 0 2.83077e-09 0.0102559 0.0534578
-0.000508544 -0 0 -8.55309e-09 0.0144758 0.0826264
-0.000475607 -0 0 2.83077e-09 0.0144603 0.055945
-0.00048232 -0 0 -8.61192e-10 0.0144635 0.0668524
-0.000573799 -0 0 2.83077e-09 0.0144983 0.055117
-0.000323799 -0 0 2.83077e-09 0.00819916 0.0483753
-0.000323799 -0 0 2.83077e-09 0.00439067 0.0618151
-0.000573799 -0 0 2.83077e-09 0.0144983 0.085265
-0.000477475 -0 0 2.83077e-09 0.0144557 0.045981
-0.000390461 -0 0 -6.07089e-09 0.00444682 0.0553409
-0.000477257 -0 0 6.35484e-09 0.0113832 0.0619901
-0.000750476 -0 0 1.25869e-08 0.0197093 0.107349
-0.000481706 -0 0 -3.52046e-10 0.0144592 0.0509564
-0.000323799 -0 0 2.83077e-09 0.0103062 0.055793
-0.00046289 -0 0 2.83077e-09 0.014456 0.0756933
-0.000495254 -0 0 8.29449e-09 0.0144655 0.0462053
-0.000373825 -0 0 2.83077e-09 0.0108125 0.051343
-0.000573799 -0 0 2.83077e-09 0.0144983 0.0721011
-0.000481413 -0 0 5.48883e-09 0.0144615 0.0403123
-0.000378721 -0 0 2.83077e-09 0.00595095 0.0527988
-0.000573799 -0 0 2.83077e-09 0.0144983 0.102255
-0.000400382 -0 0 -4.43369e-09 0.0069007 0.0287226
6.30042e-05 -0 0 2.83077e-09 -0.0026651 0.0452635
-0.000435495 -0 0 2.83077e-09 0.0097738 0.0547686
-0.000469106 -0 0 2.83077e-09 0.00982248 0.0560717
0.000125741 -0 0 2.83077e-09 -0.00412512 0.0484292
-0.000462934 -0 0 2.83077e-09 0.00751252 0.0650845
-0.000667804 -0 0 -5.24348e-09 0.0145207 0.0721987
-0.000424832 -0 0 3.74968e-09 0.00839867 0.0542887
-0.00050323 -0 0 3.74968e-09 0.0144516 0.083276
-0.00118624 -0 0 3.74968e-09 0.0289049 0.10322
-0.000578415 -0 0 3.74968e-09 0.0157659 0.0492656
-0.000412126 -0 0 3.74968e-09 0.00332497 0.054508
-0.000404368 -0 0 3.74968e-09 0.0123336 0.0578388
-0.00051215 -0 0 9.10391e-09 0.0123738 0.0910823
-0.000404368 -0 0 3.74968e-09 0.00882121 0.0543322
-0.000404368 -0 0 3.74968e-09 0.0123336 0.0539334
-0.000404368 -0 0 3.74968e-09 0.00871471 0.0561
-0.000404368 -0 0 3.74968e-09 0.00873877 0.0555488
-0.000507047 -0 0 -5.76856e-09 0.0123691 0.0897949
-0.000768395 -0 0 1.24991e-08 0.0158443 0.0859467
-0.000392008 -0 0 2.32529e-09 0.00623554 0.0343081
-0.000392008 -0 0 2.32529e-09 0.0122711 0.0456019
-0.000392008 -0 0 2.32529e-09 0.0122711 0.0526101
-0.000392008 -0 0 2.32529e-09 0.00931002 0.0517353
-0.000392008 -0 0 2.32529e-09 0.00584972 0.0423322
-0.000392008 -0 0 2.32529e-09 0.0122711 0.0527413
-0.000392008 -0 0 2.32529e-09 0.0122711 0.0619527
-0.000537575 -0 0 -2.55389e-09 0.012324 0.0721128
-0.000457688 -0 0 -3.89335e-10 0.0122959 0.0840514
-0.000474748 -0 0 -6.42487e-10 0.0123049 0.0461615
-0.000392008 -0 0 2.32529e-09 0.00542904 0.0464291
-0.000392008 -0 0 2.32529e-09 0.0122711 0.0504269
-0.000392008 -0 0 2.32529e-09 0.0122711 0.0719527
-0.000392008 -0 0 2.32529e-09 0.0122711 0.0507589
-0.000392008 -0 0 2.32529e-09 0.00858723 0.0541215
-0.000392008 -0 0 2.32529e-09 0.008138 0.0656587
-0.000551162 -0 0 2.32529e-09 0.0123354 0.0721295
-0.000392008 -0 0 2.32529e-09 0.00491105 0.0497302
-0.000482046 -0 0 -6.237e-10 0.0123049 0.0720546
-0.0006911 -0 0 2.32529e-09 0.0194784 0.0669943
-0.000360711 -0 0 2.32529e-09 0.00555064 0.0250721
-0.000347486 -0 0 2.32529e-09 0.00468455 0.0332049
-0.000388303 -0 0 2.32529e-09 0.0122163 0.0419559
0.000231651 -0 0 2.32529e-09 -0.00607035 0.0548363
-0.000304605 -0 0 2.32529e-09 0.0079979 0.0580075
-0.000170488 -0 0 2.32529e-09 0.0029905 0.0312546
0.000271837 -0 0 -1.80617e-09 -0.00468443 0.0380728
0.000268027 -0 0 1.17999e-08 -0.00618964 0.0445663
0.000261084 -0 0 1.35479e-08 -0.00598816 0.0452548
0.000181061 -0 0 2.32529e-09 -0.00862791 0.0828628
0.00036988 -0 0 2.32529e-09 -0.0119828 0.0501614
0.00027085 -0 0 1.25974e-08 -0.00556844 0.0679252
0.000181061 -0 0 2.32529e-09 -0.0060663 0.0913342
0.000276382 -0 0 -2.619e-09 -0.0119468 0.0343135
0.000274122 -0 0 2.32529e-09 -0.00934721 0.0487565
0.000322834 -0 0 1.18787e-08 -0.0061962 0.0650543
0.000181061 -0 0 2.32529e-09 -0.00699194 0.0828496
0.000181061 -0 0 2.32529e-09 -0.00716909 0.0417544
0.000339541 -0 0 -5.74703e-09 -0.00958281 0.0612099
-0.000146911 -0 0 2.32529e-09 0.00494863 0.105116
0.000181061 -0 0 2.32529e-09 -0.00365363 0.0208984
0.000288511 -0 0 2.32529e-09 -0.0119517 0.0223716
0.00027053 -0 0 6.85314e-09 -0.00629294 0.0500681
-0.000236648 -0 0 2.32529e-09 0.00532296 0.0606858
0.000181061 -0 0 2.32529e-09 -0.00860397 0.0279844
0.000244789 -0 0 -2.63329e-09 -0.00729157 0.0500972
2.38631e-06 -0 0 2.32529e-09 0.00281907 0.0794415
0.000181061 -0 0 2.32529e-09 -0.00350589 0.0501742
0.000181061 -0 0 2.32529e-09 -0.00955198 0.0625052
-2.97965e-06 -0 0 2.32529e-09 0.00210593 0.0949698
0.000181061 -0 0 2.32529e-09 -0.00830979 0.043568
0.000298899 -0 0 -5.45859e-09 -0.0119505 0.0534515
0.000181061 -0 0 2.32529e-09 -0.00190411 0.0874287
0.000181061 -0 0 2.32529e-09 -0.00374029 0.0634447
0.000317519 -0 0 2.32529e-09 -0.0119585 0.0529064
0.000181061 -0 0 2.32529e-09 -0.00312477 0.0772102
7.06293e-05 -0 0 2.32529e-09 0.00037782 0.0733328
0.000289854 -0 0 7.07188e-09 -0.00840966 0.0473801
0.000331191 -0 0 -5.55193e-09 -0.0119579 0.0599957
-0.000241041 -0 0 2.32529e-09 0.0050688 0.0806819
0.000181061 -0 0 2.32529e-09 -0.00190411 0.0167831
0.000181061 -0 0 2.32529e-09 -0.00844981 0.0195735
0.000340188 -0 0 2.32529e-09 -0.0119715 0.0361546
0.000181061 -0 0 2.32529e-09 -0.00597024 0.031775
0.000181061 -0 0 2.32529e-09 -0.00214056 0.0480441
0.000181061 -0 0 2.32529e-09 -0.00407231 0.015303
0.000181061 -0 0 2.32529e-09 -0.00520885 0.0195735
0 -0 0 0 0 0.0343214
```
Looking further, I was able to reproduce this output by running mcflirt on that specific run using SBref as a reference for the realignment.
I then tried two things:
-re-run mcflirt using the first volume of the run as reference
-re-run mcflirt using SBref as reference, but this time using another cost function: mutual info instead of the mcflirt default which is normcorr.
Here is the result, illustrated by plotting the second column of the .par file for these 3 runs of mcflirt:
1. mcflirt with SBref as reference and cost=default (normcorr) (green curve)
2. mcflirt with SBref as reference and cost=mutualinfo (red curve)
3. mcflirt with first volume as reference and cost=default (normcorr) (blue curve)

Is it something that has been observed before? Looking at the HCPpipelines code and mcflirt.sh in particular, it looks like SBref is not used directly as the reference for mcflirt (but rather the Mean of volumes 11-20) and thus a similar behavior may not happen with HCPPipeline...
jsein
July 12, 2023, 10:06pm
13
I agree, register to the first volume or to the average of the first 10 volumes as it is done I believe in the HCPPipelines.
jsein
July 12, 2023, 10:30pm
14
I don’t remember if there was any issue to use mutualinfo with mcflirt without SBref as a target for HMC. I will check back.
effigies:
ked at the code again, and we are using the sbrefs for the initial bold reference, which is used for head motion correction, but not for the final, which is used for coregistration. This needs fixing in an updated 23.2 series. That is unlikely to happen this month.
Hi @effigies thank you for this reply. If it is not using the sbref for registration, what is it using? The median volume?
We motion-correct and take the mean volume, then apply some bias-field correction.