Hi,
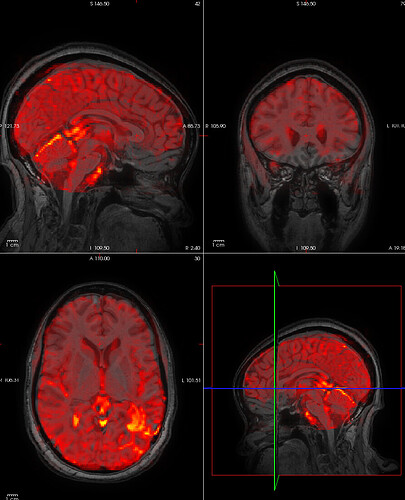
In our experiment, participants are scanned twice so there are two sessions. The participants have some type of lesion in their brain. For one participant, we are experiencing poor anatomical and functional coregistration for the second session only for all functional runs except one. I tried manually setting the origin for each scan, so there is good initial overlap between functional and anatomical images. Here is one example of a processed functional overlaid on the processed T1w:
We are using fmriprep 20.2.1 and here is the command used:
singularity run --cleanenv \
-B /cm/shared/apps/freesurfer/6.0.0:/work_dir \
-B /scratch/global/jheffernan/experiments/tdcs/templateflow:/opt/templateflow \
/scratch/global/jheffernan/experiments/tdcs/singularity_images/fmriprep-v20.2.1.simg \
/scratch/global/jheffernan/experiments/tdcs/bids /scratch/global/jheffernan/experiments/tdcs/derivatives/fmriprep-v20.2.1/sessions-2_manualorigin participant \
--participant_label mcwa002 \
--nprocs 16 \
--omp-nthreads 4 \
--ignore slicetiming \
--output-spaces T1w \
--random-seed 1234567890 \
--fs-license-file /work_dir/license.txt \
--fs-no-reconall \
--work-dir /scratch/global/jheffernan/experiments/tdcs/tmp/fmriprep_mcwa002_ALL_sessions-2_IDvxTZ \
--stop-on-first-crash
We did not include a lesion mask, because we are only using T1w output-space, so I don’t think it would help. We use --fs-no-reconall, because the brain mask refinement with freesurfer tends to exclude the lesioned areas of the brain. I tried --force-no-bbr option and the results did not look good either. The functional scanning dimensions also changed between sessions, but not much so I do not think it should be a big problem. Here are the parameters for both sessions:
sub-mcwa002_ses-1_task-bh_run-1_bold.nii.gz int16 [128, 128, 72, 330] 1.62x1.62x2.00x0.80
sub-mcwa002_ses-1_task-rest_run-1_bold.nii.gz int16 [128, 128, 72, 378] 1.62x1.62x2.00x0.80
sub-mcwa002_ses-1_task-rest_run-2_bold.nii.gz int16 [128, 128, 72, 378] 1.62x1.62x2.00x0.80
sub-mcwa002_ses-1_task-rest_run-3_bold.nii.gz int16 [128, 128, 72, 378] 1.62x1.62x2.00x0.80
sub-mcwa002_ses-1_task-SDTD_run-1_bold.nii.gz int16 [128, 128, 72, 378] 1.62x1.62x2.00x0.80
sub-mcwa002_ses-1_task-SDTD_run-2_bold.nii.gz int16 [128, 128, 72, 378] 1.62x1.62x2.00x0.80
sub-mcwa002_ses-2_task-bh_run-1_bold.nii.gz int16 [128, 128, 68, 176] 1.62x1.62x2.00x1.50
sub-mcwa002_ses-2_task-rest_run-1_bold.nii.gz int16 [128, 128, 68, 201] 1.62x1.62x2.00x1.50
sub-mcwa002_ses-2_task-rest_run-2_bold.nii.gz int16 [128, 128, 68, 201] 1.62x1.62x2.00x1.50
sub-mcwa002_ses-2_task-rest_run-3_bold.nii.gz int16 [128, 128, 68, 201] 1.62x1.62x2.00x1.50
sub-mcwa002_ses-2_task-rest_run-4_bold.nii.gz int16 [128, 128, 68, 201] 1.62x1.62x2.00x1.50
sub-mcwa002_ses-2_task-SDTD_run-1_bold.nii.gz int16 [128, 128, 68, 201] 1.62x1.62x2.00x1.50
sub-mcwa002_ses-2_task-SDTD_run-2_bold.nii.gz int16 [128, 128, 68, 201] 1.62x1.62x2.00x1.50