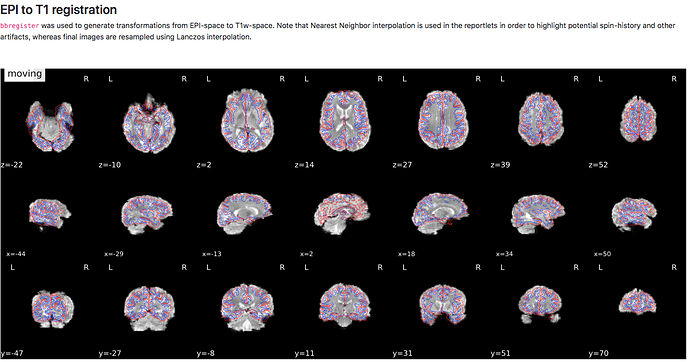
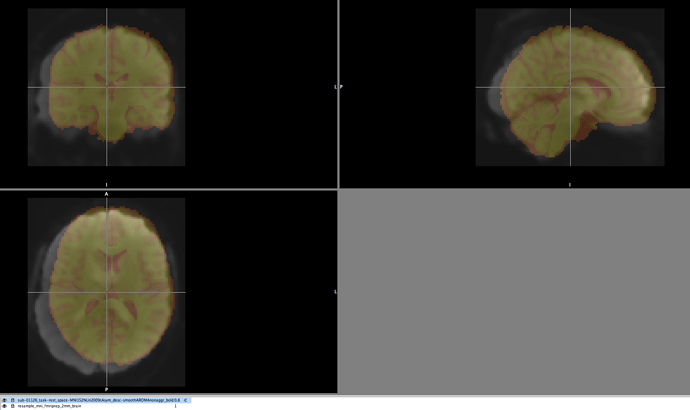
Hi,
When running fmriprep, it seems as though both the EPI to T1 registration and the EPI to MNI registrations performed poorly.
This is the command that I used:
singularity run --cleanenv -B “${myproject}”:"${myproject}" “${myproject}/fmriprep-1.3.0.post2.simg” “${myproject}/PNC_bids_full” “${myproject}/output” participant --participant-label “${SUBJECT}” --fs-license-file “${myproject}/license.txt” --use-aroma
I was wondering if you have seen something like this before and if you have any recommendations on how to improve the registration.
Thank you
Here is an image demonstrating the poor registration to MNI
With the red image being the MNI version used by fmriprep, while the withe being a participant’s bold image
Hi @neuro_kf,
Could you share the full report (please make sure the sub-${SUBJECT}/figures folder is included).
It seems to me that this may not a problem of registration between functional and anatomical, rather than a problem in susceptibility distortion correction. The BOLD images seem odd.
Here is a link to the output for this subject and the html file.
Thank you for the quick reply
Hi @neuro_kf, I can see that there wasn’t any information to perform susceptibility distortion correction, nor you used the --use-syn-sdc flag.
One possibility is that there is an spurious X-axis flip in either your T1w or the functional scan.
Another possibility is that your BOLD sequence is not correctly shimmed because it seems to have a quite acute, low frequency distortion that is too much for the typical distortion derived from susceptibility inhomogeneity. I’d recommend to revise your protocol and consluting with your technician. That said, you may salvage these data using the --use-syn-sdc option. In other words, I honestly doubt you can get any better registration with these data, there’s something odd on the functional scan.
The spatial normalization of the T1 looks pretty good, so the misalignment you see w.r.t. MNI (what MNI is this?) could be derived from your viewer misinterpreting x-form headers of the nifti file.
Finally, please remove the anat/ folder from the OSF as those T1w images are not defaced (or please double check you can share them otherwise).
Thank you for the advice.
We can close the issue there.
If you had time/resources, it would be great to have your feedback confirming whether my intuitions were correct, and to what extent using --use-syn-sdc alleviated the issue.