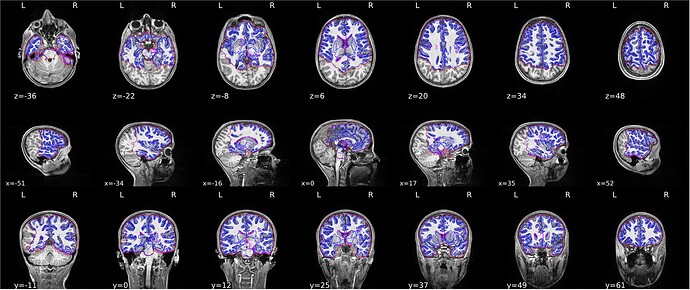
Summary of what happened:
Despite observed correct segmentation from freesurfer recon, during QC of the fmriprept HTML report we found large portions of what and gray matter were not segmented for a handful of particpants. recon was ran before running fmri prep.
Command used (and if a helper script was used, a link to the helper script or the command generated):
singularity_cmd = f"singularity run \
--cleanenv \
/home/data/madlab/singularity-images/nipreps_fmriprep_20.2.3.sif \
{output_dir}/fmriprep_dset/ \
{output_dir}/fmriprep_derivatives/ participant \
-w /scratch/madlab/fmriprep/emuR01_work \
--participant_label {' '.join(subjects)} \
--skull-strip-template MNIPediatricAsym:cohort-5 \
--output-spaces MNIPediatricAsym:cohort-5:res-2 \
--fs-license-file=/scratch/madlab/license.txt \
--skip-bids-validation \
--bids-database-dir {output_dir}/bids_layout \
--clean-workdir \
--nthreads 40 \
--omp-nthreads 40"
Version:
20.2.3
Environment (Docker, Singularity, custom installation):
Data formatted according to a validatable standard? Please provide the output of the validator:
BIDS standard
Relevant log outputs (up to 20 lines):
Screenshots / relevant information:
From fmriprep: