Summary of what happened:
Dear all,
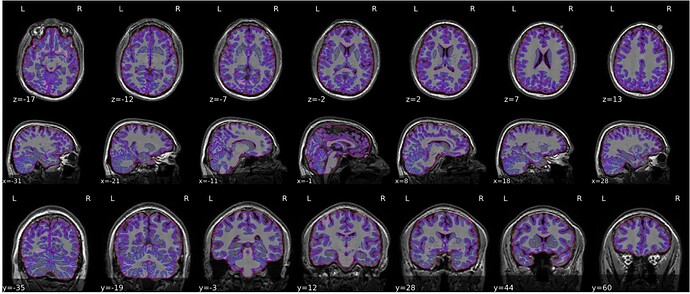
We used fmriprep-20.2.7 to preprocess our data, but we are facing issues with poor skull-strip which very consistent across subjects (see the attached image, x = 18; y = 44). For context, our MRI data were acquired by GE scanner, and the T1w contrast look fine and there was no intensity in-homogeneity issue.
Command used (and if a helper script was used, a link to the helper script or the command generated):
> usr/local/miniconda/bin/fmriprep /bids /output participant
--skip_bids_validation --participant_label CAMH036 --nprocs 4 --omp-nthreads 2
-vv --output-spaces T1w MNI152NLin2009cAsym fsaverage fsaverage5
--bold2t1w-init register --bold2t1w-dof 6 --use-aroma
--aroma-melodic-dimensionality -200 --use-syn-sdc
--fs-license-file /license/license.txt --no-submm-recon -w /work
Version:
20.2.7 LTS
Environment (Docker, Singularity, custom installation):
Anaconda Python
Data formatted according to a validatable standard? Please provide the output of the validator:
data were organized following BIDS standard.
sub-CAMH036/
├── ses-posttx
│ ├── anat
│ │ ├── sub-CAMH036_ses-posttx_T1w.json
│ │ └── sub-CAMH036_ses-posttx_T1w.nii.gz
│ └── func
│ ├── sub-CAMH036_ses-posttx_task-rest_bold.json
│ └── sub-CAMH036_ses-posttx_task-rest_bold.nii.gz
└── ses-pretx
├── anat
│ ├── sub-CAMH036_ses-pretx_T1w.json
│ └── sub-CAMH036_ses-pretx_T1w.nii.gz
└── func
├── sub-CAMH036_ses-pretx_task-rest_bold.json
└── sub-CAMH036_ses-pretx_task-rest_bold.nii.gz
Relevant log outputs (up to 20 lines):
TASK ATTEMPT 1
============================
docker is /usr/bin/docker
[ INFO ] Descriptor from execution saved to cache for future publishing as descriptor_fmriprep-(BIDS-app)_2022-10-21T21:52:28.712050.json
[ INFO ] Input: {'bids_dir': '/bids', 'output_dir': '/output', 'work_dir': '/work', 'analysis_level': 'participant', 'fs_license_file': '/license/license.txt', 'nprocs': 4, 'omp_nthreads': 2, 'verbosity': '-vv', 'use_aroma': True, 'use_syn_sdc': True, 'skip_bids_validation': True, 'output_spaces': ['T1w', 'MNI152NLin2009cAsym', 'fsaverage', 'fsaverage5'], 'participant_label': ['CAMH036']}
[ INFO ] Running: chmod 755 /tmp/nxf.Oxw0wp6DkJ/temp-1061311154703-1666403548720.localExec.boshjob.sh
[ INFO ] Running: singularity exec --cleanenv -B /mnt/tigrlab/projects/ttan/ThirtyFourD/THREED_BIDS:/bids -B /mnt/tigrlab/projects/ttan/ThirtyFourD/in_progress:/output -B /mnt/tigrlab/quarantine/freesurfer/6.0.0/build:/license -B /tmp/nxf.Oxw0wp6DkJ/work:/work -B /tmp/nxf.Oxw0wp6DkJ:/tmp/nxf.Oxw0wp6DkJ -W /tmp/nxf.Oxw0wp6DkJ /archive/code/containers/FMRIPREP/nipreps_fmriprep_20.2.7-2022-01-24-5df135ac568c.simg /tmp/nxf.Oxw0wp6DkJ/temp-1061311154703-1666403548720.localExec.boshjob.sh
/usr/local/miniconda/lib/python3.7/site-packages/bids/config.py:40: FutureWarning: Setting 'extension_initial_dot' will be removed in pybids 0.16.
FutureWarning)
221022-01:55:46,988 cli INFO:
Telemetry system to collect crashes and errors is enabled - thanks for your feedback!. Use option ``--notrack`` to opt out.
You are using fMRIPrep-20.2.7, and a newer version of fMRIPrep is available: 22.0.2.
Please check out our documentation about how and when to upgrade:
https://fmriprep.readthedocs.io/en/latest/faq.html#upgrading
221022-01:56:01,226 nipype.workflow WARNING:
Previous output generated by version 0+unknown found.
221022-01:56:04,188 nipype.workflow IMPORTANT:
Running fMRIPREP version 20.2.7:
* BIDS dataset path: /bids.
Screenshots / relevant information:
We have tried solutions to address this major skullstrip problem in our dataset
- Use
--skull-strip-t1woption. The output did not improve. - Use
Roburst Brain Extraction (ROBEX). This also did not improve the skull-strip - defaced T1w.
- We are currently trying to change parameters in the ANTS brain extraction to see if it improve the skull-strip.
- Would running with newer fmriprep version fix our current problem?
We would really appreciate your expertise and comments on how to improve this skull-strip issue.
Best,
Thomas