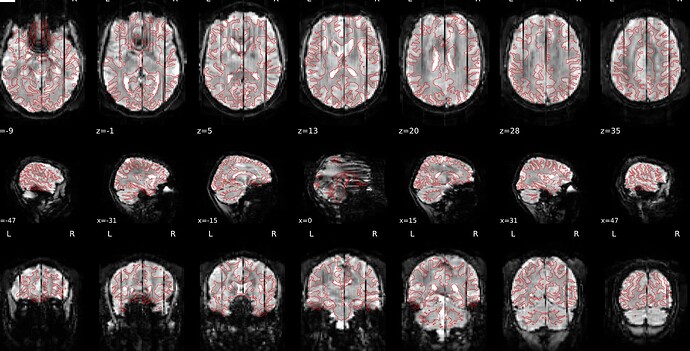
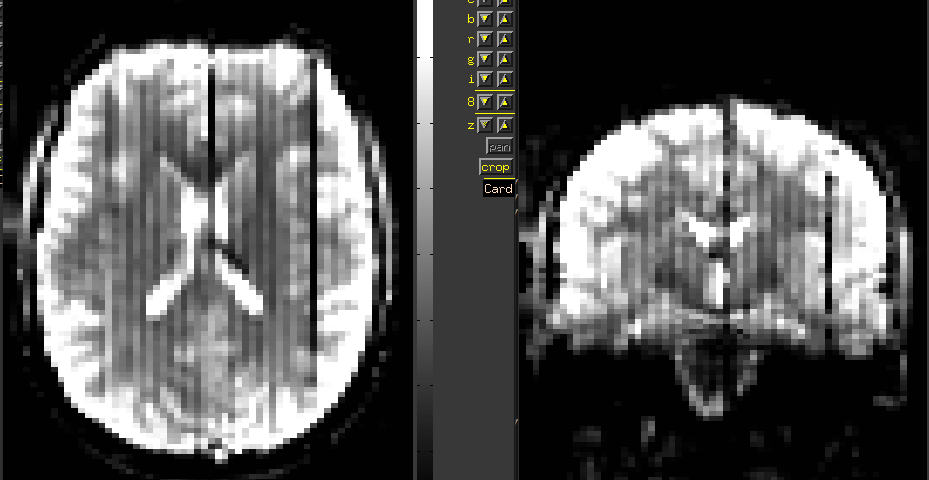
Summary of what happened:
Hi all,
I am a first-time user of fmriprep, and have observed an issue with several of my participants. An example co-registration image is attached — you can see a number of dark streaks, running in the posterior-anterior direction. I’ve observed this in about 20 of the 70 participants I’ve run fmriprep on, but haven’t been able to find anything else in the QA report that corresponds with this (these participants don’t have excess motion, for example). Do any of you have insight into what might be going wrong here?
Command used (and if a helper script was used, a link to the helper script or the command generated):
fmriprep-docker --fs-license-file ../Software/license.txt stagingareas/${user} /home/${user}
Version:
23.2.1
Environment (Docker, Singularity / Apptainer, custom installation):
Singularity
Data formatted according to a validatable standard? Please provide the output of the validator:
Sidecars were written using dcm2niix -- errors did not prevent fmriprep from running
error: SIDECAR_KEY_REQUIRED (TaskName)
warning: README_FILE_MISSING
warning: TOO_FEW_AUTHORS
warning: JSON_KEY_RECOMMENDED (DatasetType)
warning: JSON_KEY_RECOMMENDED (License)
warning: JSON_KEY_RECOMMENDED (GeneratedBy)
warning: JSON_KEY_RECOMMENDED (SourceDatasets)
warning: NO_AUTHORS (Authors)
warning: SIDECAR_KEY_RECOMMENDED (ReceiveCoilName)
warning: SIDECAR_KEY_RECOMMENDED (ReceiveCoilActiveElements)
warning: SIDECAR_KEY_RECOMMENDED (GradientSetType)
warning: SIDECAR_KEY_RECOMMENDED (MRTransmitCoilSequence)
warning: SIDECAR_KEY_RECOMMENDED (MatrixCoilMode)
warning: SIDECAR_KEY_RECOMMENDED (CoilCombinationMethod)
warning: SIDECAR_KEY_RECOMMENDED (PulseSequenceType)
warning: SIDECAR_KEY_RECOMMENDED (SequenceName)
warning: SIDECAR_KEY_RECOMMENDED (PulseSequenceDetails)
warning: SIDECAR_KEY_RECOMMENDED (MTState)
warning: SIDECAR_KEY_RECOMMENDED (SpoilingState)
warning: SIDECAR_KEY_RECOMMENDED (NumberShots)
warning: SIDECAR_KEY_RECOMMENDED (ParallelReductionFactorOutOfPlane)
warning: SIDECAR_KEY_RECOMMENDED (ParallelAcquisitionTechnique)
warning: SIDECAR_KEY_RECOMMENDED (PartialFourier)
warning: SIDECAR_KEY_RECOMMENDED (PartialFourierDirection)
warning: SIDECAR_KEY_RECOMMENDED (EffectiveEchoSpacing)
warning: SIDECAR_KEY_RECOMMENDED (MixingTime)
warning: SIDECAR_KEY_RECOMMENDED (PhaseEncodingDirection)
warning: SIDECAR_KEY_RECOMMENDED (TotalReadoutTime)
warning: SIDECAR_KEY_RECOMMENDED (DwellTime)
warning: SIDECAR_KEY_RECOMMENDED (MultibandAccelerationFactor)
warning: SIDECAR_KEY_RECOMMENDED (InstitutionAddress)
warning: SIDECAR_KEY_RECOMMENDED (InstitutionalDepartmentName)
warning: SIDECAR_KEY_RECOMMENDED (TaskName)
warning: SIDECAR_KEY_RECOMMENDED (NumberOfVolumesDiscardedByScanner)
warning: SIDECAR_KEY_RECOMMENDED (NumberOfVolumesDiscardedByUser)
warning: SIDECAR_KEY_RECOMMENDED (DelayTime)
warning: SIDECAR_KEY_RECOMMENDED (AcquisitionDuration)
warning: SIDECAR_KEY_RECOMMENDED (DelayAfterTrigger)
warning: SIDECAR_KEY_RECOMMENDED (Instructions)
warning: SIDECAR_KEY_RECOMMENDED (TaskDescription)
warning: SIDECAR_KEY_RECOMMENDED (CogAtlasID)
warning: SIDECAR_KEY_RECOMMENDED (CogPOID)
warning: SIDECAR_KEY_RECOMMENDED (ParallelReductionFactorInPlane)
warning: SIDECAR_KEY_RECOMMENDED (InversionTime)
warning: SIDECAR_KEY_RECOMMENDED (SliceEncodingDirection)
warning: EVENTS_TSV_MISSING
Relevant log outputs (up to 20 lines):
anat_only = false
aroma_err_on_warn = false
aroma_melodic_dim = 0
bold2t1w_dof = 6
bold2t1w_init = "register"
cifti_output = false
fmap_bspline = false
force_syn = false
hires = true
ignore = []
level = "full"
longitudinal = false
run_msmsulc = true
medial_surface_nan = false
project_goodvoxels = false
regressors_all_comps = false
regressors_dvars_th = 1.5
regressors_fd_th = 0.5
run_reconall = true
skull_strip_fixed_seed = false
skull_strip_template = "OASIS30ANTs"
skull_strip_t1w = "force"
slice_time_ref = 0.5
spaces = "MNI152NLin2009cAsym:res-native"
use_aroma = false
use_syn_sdc = false
me_t2s_fit_method = "curvefit"