Summary of what happened:
Hi there,
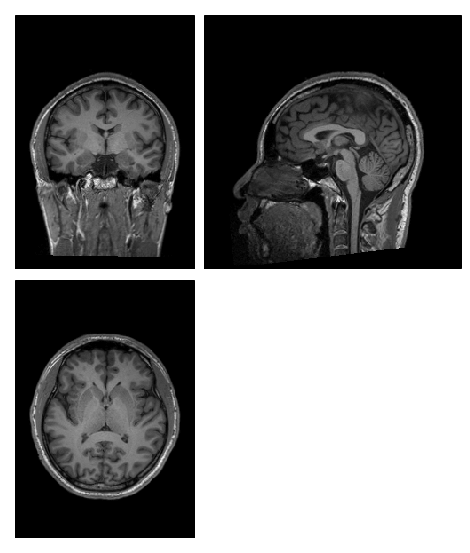
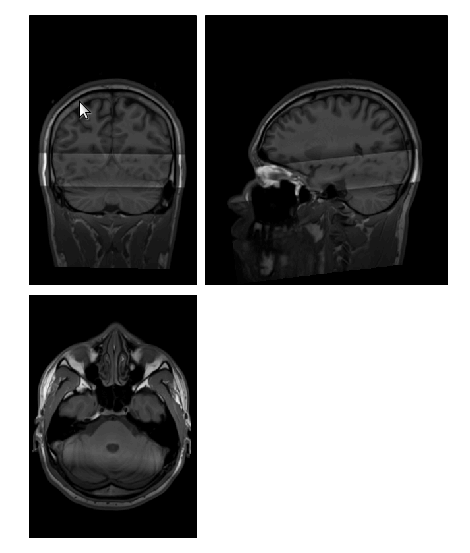
I’m having something strange happen to my preprocessed anatomical images. They go into fMRIPrep looking perfectly normal (raw bids file, first image), and they come out with a strange, light coloured bar across the brain along with a lot of shadowing (as you can see in the second image).
Can anyone help me understand what’s going on and why the images are coming out this way? Is there anything I can do to solve the issue? I was originally putting into into MNI template space, but was running into the same problem. Any input on this issue would be greatly appreciated.
Cheers,
Emily
Command used (and if a helper script was used, a link to the helper script or the command generated):
Below is the python script I am using to run fMRIPrep:
def run_fmriprep(input_dir, output_dir, subject):
input_path = os.path.join(input_dir, 'BIDS_ya')
output_path = os.path.join(output_dir, 'fmriprep')
license_file = os.path.join(output_dir, 'license.txt')
freesurfer_path = os.path.join(derv_dir, 'freesurfer_ya')
# Get the current user's UID and GID
uid = os.getuid()
gid = os.getgid()
docker_command = [
'docker', 'run', '--rm',
'--user', f'{uid}:{gid}', # Add the --user flag with UID and GID
'-v', f'{input_path}:/data:ro',
'-v', f'{output_path}:/out',
'-v', f'{license_file}:/opt/freesurfer/license.txt:ro',
'-v', f'{freesurfer_path}:/freesurfer_subjects',
'-v', f'{output_dir}:/work',
'nipreps/fmriprep:latest',
'/data', '/out', 'participant',
'--participant-label', subject.replace('sub-', ''),
'--skip-bids-validation',
'--use-aroma',
'--md-only-boilerplate',
'--output-spaces', 'func',
'--ignore', 'slicetiming',
'--dummy-scans', '4',
'--n_cpus', '5',
'--skull-strip-t1w', 'force',
'--me-output-echos',
'--low-mem',
'-w', '/work'
]
Version:
Environment (Docker, Singularity / Apptainer, custom installation):
Docker (running Docker through Python)
Data formatted according to a validatable standard? Please provide the output of the validator:
PASTE VALIDATOR OUTPUT HERE
Relevant log outputs (up to 20 lines):
fMRIPrep ran successfully.