Hi Marcel,
Thank you for your help so far - really appreciate it. In summary, it works but only if I manually delete the radiology report files.
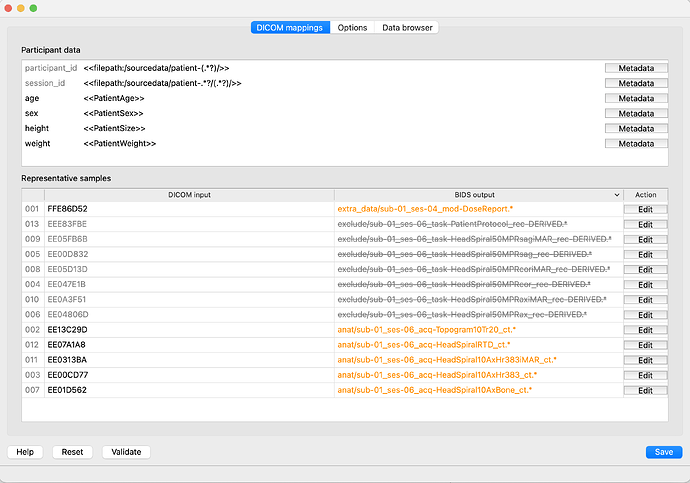
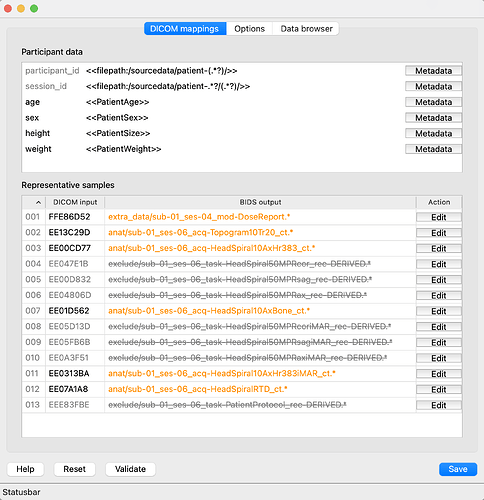
Interestingly, at first, it picks up only ses-4 and ses-6 (see screenshot). It appears that the 5 radiology report files (which are missing ‘SeriesDescription’) are blocking it from creating subfolders:
2025-03-19 18:12:06 - ERROR | Cannot create subfolders, aborting dicomsort()…
2025-03-19 18:12:06 - ERROR | Missing ‘SeriesDescription’ DICOM field specified in the ‘{SeriesNumber:03d}-{SeriesDescription}’ folder/naming scheme, cannot find a safe name for: /private/var/folders/ … sourcedata/patient-01/DICOM/0000AC37/AA740734/AAB9BE77/00007A3B/FFA7A389.
Does the above suggest that it’s not the DS_Store files which are causing the problem - instead the report files?
It also gives a BIDScoiner version difference, but I don’t think this is a problem: /Users/user/.bidscoin/4.5.2.dev0/templates/bidsmap_dccn.yaml was created with version 4.5.2.dev, but this is version 4.5.2.dev0. This is normally OK but check the Changelog — BIDScoin 4.5.2.dev documentation
The bidseditor GUI does not seem to have full functionality, e.g. sliding the column to expand the width under ‘Representative Sample’ as before. Is this intentional because it is a developer version or an error at my end?
So, I manually deleted the files which gave the error above, and re-ran the code. It was then able to work, including bidscoiner, which gave the following output (below).
I think the BIDS output structure looks correct, however, I had to manually delete the files in order for this code to work: bidsmapper sourcedata BIDS_new -s -n ‘patient-’ -m ‘*’
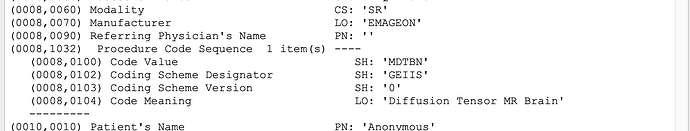
I have checked the radiology reports and they all have the dicom tag (0008,0104) CodeMeaning with value ‘Radiology Report’. Do you think the block comes from the radiology reports and is there a way to exclude them based on this in order to automate the process?
sub-01
├── ses-01
│ ├── extra_data
│ │ ├── sub-01_ses-01_mod-MagImages_echo-1_GR.json
│ │ ├── sub-01_ses-01_mod-MagImages_echo-1_GR.nii.gz
│ │ ├── sub-01_ses-01_mod-PhaImages_echo-1_part-phase_GR.json
│ │ ├── sub-01_ses-01_mod-PhaImages_echo-1_part-phase_GR.nii.gz
│ │ ├── sub-01_ses-01_mod-SWIImages_echo-1_GR.json
│ │ ├── sub-01_ses-01_mod-SWIImages_echo-1_GR.nii.gz
│ │ ├── sub-01_ses-01_mod-t1mp2ragesagp3isoINV1_echo-1_GRIR.json
│ │ ├── sub-01_ses-01_mod-t1mp2ragesagp3isoINV1_echo-1_GRIR.nii.gz
│ │ ├── sub-01_ses-01_mod-t1mp2ragesagp3isoINV2_echo-1_GRIR.json
│ │ ├── sub-01_ses-01_mod-t1mp2ragesagp3isoINV2_echo-1_GRIR.nii.gz
│ │ ├── sub-01_ses-01_mod-t1mpragesagp2isopost_echo-1_GRIR.json
│ │ ├── sub-01_ses-01_mod-t1mpragesagp2isopost_echo-1_GRIR.nii.gz
│ │ ├── sub-01_ses-01_mod-t1mpragesagp2isopre_echo-1_GRIR.json
│ │ ├── sub-01_ses-01_mod-t1mpragesagp2isopre_echo-1_GRIR.nii.gz
│ │ ├── sub-01_ses-01_mod-t2spcflairsag1mmiso_echo-1_SEIR.json
│ │ ├── sub-01_ses-01_mod-t2spcflairsag1mmiso_echo-1_SEIR.nii.gz
│ │ ├── sub-01_ses-01_mod-t2tsecor448_echo-1_SE.json
│ │ ├── sub-01_ses-01_mod-t2tsecor448_echo-1_SE.nii.gz
│ │ ├── sub-01_ses-01_mod-t2tsetra448_echo-1_SE.json
│ │ └── sub-01_ses-01_mod-t2tsetra448_echo-1_SE.nii.gz
│ └── sub-01_ses-01_scans.tsv
├── ses-02
│ ├── extra_data
│ │ ├── sub-01_ses-02_mod-pCASLBL1800PLD1500con_echo-1_EP.json
│ │ ├── sub-01_ses-02_mod-pCASLBL1800PLD1500con_echo-1_EP.nii.gz
│ │ ├── sub-01_ses-02_mod-pCASLBL1800PLD1500diff_echo-1_EP.json
│ │ ├── sub-01_ses-02_mod-pCASLBL1800PLD1500diff_echo-1_EP.nii.gz
│ │ ├── sub-01_ses-02_mod-pCASLBL1800PLD1500lab_echo-1_EP.json
│ │ ├── sub-01_ses-02_mod-pCASLBL1800PLD1500lab_echo-1_EP.nii.gz
│ │ ├── sub-01_ses-02_mod-ssTE00TI1700_echo-1_EP.json
│ │ └── sub-01_ses-02_mod-ssTE00TI1700_echo-1_EP.nii.gz
│ └── sub-01_ses-02_scans.tsv
├── ses-03
│ ├── dwi
│ │ ├── sub-01_ses-03_acq-DTI2x2x22Multishell20ch2020fftscale_sbref.json
│ │ ├── sub-01_ses-03_acq-DTI2x2x22Multishell20ch2020fftscale_sbref.nii.gz
│ │ ├── sub-01_ses-03_acq-DTI2x2x22Multishell20ch2020negpefftscale_sbref.json
│ │ ├── sub-01_ses-03_acq-DTI2x2x22Multishell20ch2020negpefftscale_sbref.nii.gz
│ │ ├── sub-01_ses-03_run-1_dwi.bval
│ │ ├── sub-01_ses-03_run-1_dwi.bvec
│ │ ├── sub-01_ses-03_run-1_dwi.json
│ │ ├── sub-01_ses-03_run-1_dwi.nii.gz
│ │ ├── sub-01_ses-03_run-2_dwi.bval
│ │ ├── sub-01_ses-03_run-2_dwi.bvec
│ │ ├── sub-01_ses-03_run-2_dwi.json
│ │ └── sub-01_ses-03_run-2_dwi.nii.gz
│ └── sub-01_ses-03_scans.tsv
├── ses-04
│ ├── anat
│ │ ├── sub-01_ses-04_acq-DEHeadAngio10Hr383F08_ct.json
│ │ ├── sub-01_ses-04_acq-DEHeadAngio10Hr383F08_ct.nii.gz
│ │ ├── sub-01_ses-04_acq-DEHeadAngio10Hr383F08_ct_ROI1.nii.gz
│ │ ├── sub-01_ses-04_acq-DEPPDEHeadAngio10Qr403A80kV_ct.json
│ │ ├── sub-01_ses-04_acq-DEPPDEHeadAngio10Qr403A80kV_ct.nii.gz
│ │ ├── sub-01_ses-04_acq-DEPPDEHeadAngio10Qr403BSn150kV_ct.json
│ │ ├── sub-01_ses-04_acq-DEPPDEHeadAngio10Qr403BSn150kV_ct.nii.gz
│ │ ├── sub-01_ses-04_acq-Monitoring100Br36_ct.json
│ │ ├── sub-01_ses-04_acq-Monitoring100Br36_ct.nii.gz
│ │ ├── sub-01_ses-04_acq-Monitoring100Br36_ct_ROI1.nii.gz
│ │ ├── sub-01_ses-04_acq-PreMonitoring100Br36_ct.json
│ │ ├── sub-01_ses-04_acq-PreMonitoring100Br36_ct.nii.gz
│ │ ├── sub-01_ses-04_acq-PreMonitoring100Br36_ct_ROI1.nii.gz
│ │ ├── sub-01_ses-04_acq-Topogram10Tr20_ct.json
│ │ ├── sub-01_ses-04_acq-Topogram10Tr20_ct.nii.gz
│ │ └── sub-01_ses-04_acq-Topogram10Tr20_ct_ROI1.nii.gz
│ ├── extra_data
│ └── sub-01_ses-04_scans.tsv
├── ses-05
│ ├── extra_data
│ │ ├── sub-01_ses-05_run-10_mod-OrthoSpine.json
│ │ ├── sub-01_ses-05_run-10_mod-OrthoSpine.nii.gz
│ │ ├── sub-01_ses-05_run-11_mod-OrthoSpine.json
│ │ ├── sub-01_ses-05_run-11_mod-OrthoSpine.nii.gz
│ │ ├── sub-01_ses-05_run-12_mod-OrthoSpine.json
│ │ ├── sub-01_ses-05_run-12_mod-OrthoSpine.nii.gz
│ │ ├── sub-01_ses-05_run-1_mod-OrthoSpine.json
│ │ ├── sub-01_ses-05_run-1_mod-OrthoSpine.nii.gz
│ │ ├── sub-01_ses-05_run-2_mod-OrthoSpine.json
│ │ ├── sub-01_ses-05_run-2_mod-OrthoSpine.nii.gz
│ │ ├── sub-01_ses-05_run-3_mod-OrthoSpine.json
│ │ ├── sub-01_ses-05_run-3_mod-OrthoSpine.nii.gz
│ │ ├── sub-01_ses-05_run-4_mod-OrthoSpine.json
│ │ ├── sub-01_ses-05_run-4_mod-OrthoSpine.nii.gz
│ │ ├── sub-01_ses-05_run-5_mod-OrthoSpine.json
│ │ ├── sub-01_ses-05_run-5_mod-OrthoSpine.nii.gz
│ │ ├── sub-01_ses-05_run-6_mod-OrthoSpine.json
│ │ ├── sub-01_ses-05_run-6_mod-OrthoSpine.nii.gz
│ │ ├── sub-01_ses-05_run-7_mod-OrthoSpine.json
│ │ ├── sub-01_ses-05_run-7_mod-OrthoSpine.nii.gz
│ │ ├── sub-01_ses-05_run-8_mod-OrthoSpine.json
│ │ ├── sub-01_ses-05_run-8_mod-OrthoSpine.nii.gz
│ │ ├── sub-01_ses-05_run-9_mod-OrthoSpine.json
│ │ └── sub-01_ses-05_run-9_mod-OrthoSpine.nii.gz
│ └── sub-01_ses-05_scans.tsv
└── ses-06
├── anat
│ ├── sub-01_ses-06_acq-HeadSpiral10AxBone_ct.json
│ ├── sub-01_ses-06_acq-HeadSpiral10AxBone_ct.nii.gz
│ ├── sub-01_ses-06_acq-HeadSpiral10AxHr383_ct.json
│ ├── sub-01_ses-06_acq-HeadSpiral10AxHr383_ct.nii.gz
│ ├── sub-01_ses-06_acq-HeadSpiral10AxHr383iMAR_ct.json
│ ├── sub-01_ses-06_acq-HeadSpiral10AxHr383iMAR_ct.nii.gz
│ ├── sub-01_ses-06_acq-HeadSpiralRTD_ct.json
│ ├── sub-01_ses-06_acq-HeadSpiralRTD_ct.nii.gz
│ ├── sub-01_ses-06_acq-Topogram10Tr20_ct.json
│ └── sub-01_ses-06_acq-Topogram10Tr20_ct.nii.gz
├── extra_data
└── sub-01_ses-06_scans.tsv