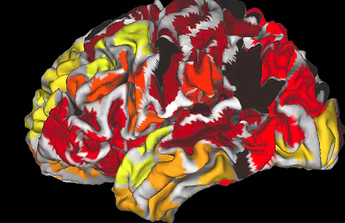
Dear All,
I am having some difficulty in using the Gordon parcellation (https://sites.wustl.edu/petersenschlaggarlab/parcels-19cwpgu/) for surface based connectivity (specifically the 164K surface version)
If I view this parcellation using connectome workbench it looks as one would expect:
However if I view it using nilearn:
from nilearn import plotting, surface, datasets
parcellation = surface.load_surf_data(atlas_l_path)
fsaverage = datasets.fetch_surf_fsaverage(mesh='fsaverage')
plotting.plot_surf_roi(fsaverage['pial_left'], roi_map=parcellation,
hemi='left', view='lateral',
bg_map=fsaverage['sulc_left'], bg_on_data=True,
darkness=.5)
The rois are broken up and shuffled across the cortex,

This is reflected in the fact that when I try and build a connectivity matrix the expected structure is not seen.
def surface_conn(sub_id, denoised_path):
atlas_l_path = f'/home/k1201869/glu_rs/data/atlas_data/gordon/notworking'
atlas_r_path = f'/home/k1201869/glu_rs/data/atlas_data/gordon/notworking'
surf_image_l_path = f'{denoised_path}/sub-{sub_id}/regress/lh.sub-{sub_id}_residualised.mgz'
surf_image_r_path = f'{denoised_path}/sub-{sub_id}/regress/rh.sub-{sub_id}_residualised.mgz'
surf_image_l = surface.load_surf_data(surf_image_l_path)
atlas_image_l = surface.load_surf_data(atlas_l_path)
surf_image_r = surface.load_surf_data(surf_image_r_path)
atlas_image_r = surface.load_surf_data(atlas_r_path)
l_roi_timecourses = []
# We don't include the first roi as this is background
for i in np.unique(atlas_image_l)[1:]:
roi_idxs = np.where(atlas_image_l==i)
l_roi_timecourses.append(np.mean(surf_image_l[roi_idxs], axis=0))
r_roi_timecourses = []
for i in np.unique(atlas_image_r)[1:]:
roi_idxs = np.where(atlas_image_r==i)
r_roi_timecourses.append(np.mean(surf_image_r[roi_idxs], axis=0))
bilat_timecourses = np.array(l_roi_timecourses + r_roi_timecourses)
connectivity_matrix = np.corrcoef(bilat_timecourses)
return connectivity_matrix
So it seems as though when loading the parcellation with nilearn the vertices are getting shuffled, I wonder if anyone has any advice about how to fix this?
Thanks!
Rob
Edit: I think I just need to convert from HCP to FS space following the steps here: https://figshare.com/articles/HCP-MMP1_0_projected_on_fsaverage/3498446