Hello everyone,
I’m new to both neuroimaging and this site. My name is Szymon, and I’m an MD currently pursuing a PhD in whole-brain modeling. I would be extremely grateful for any assistance with the following problem:
I am currently working with data from the UCLA Consortium for Neuropsychiatric Phenomics LA5c Study that has been preprocessed using fMRIPrep. I have obtained resting-state data for several healthy subjects. My objective is to parcelate signals using the Schaefer 100 atlas, regress out motion parameters (I was trying with three translational and three rotational) and to apply a filter to the signals within the 0.01-0.1 Hz frequency range. For these tasks, I am using Nilearn and have written below Python code. As a preliminary step, I have eliminated subjects whose data show translations (X, Y, Z) greater than 1.5 mm and rotations (RotX, RotY, RotZ) exceeding 1.5 degrees.
fmri_filename = f’{subject}_task-rest_bold_space-MNI152NLin2009cAsym_preproc.nii’
atlas_filename = 'Schaefer2018_100Parcels_7Networks_order_FSLMNI152_2mm.nii'
confounds_filename = f'{subject}_task-rest_bold_confounds.tsv'
confounds = pd.read_csv(confounds_filename, sep='\t')
motion_parameters = confounds[['X', 'Y', 'Z', 'RotX', 'RotY', 'RotZ']]
masker = input_data.NiftiLabelsMasker(labels_img=atlas_filename, standardize='zscore_sample',
memory='nilearn_cache', verbose=5)
time_series = masker.fit_transform(fmri_filename, confounds=motion_parameters.values)
filtered_time_series = signal.clean(time_series, detrend=True, standardize='zscore_sample', low_pass=0.1, high_pass=0.01, t_r=2.0)
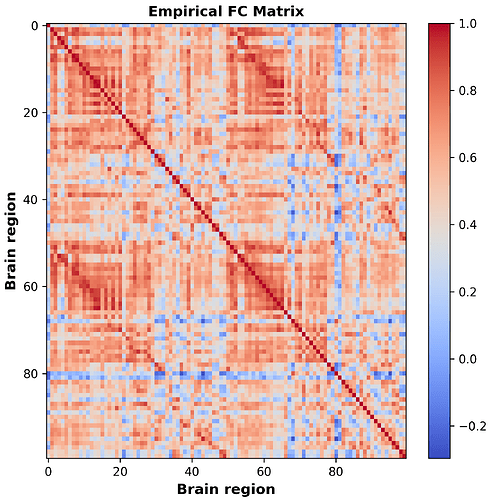
Everything is functioning, but the output Functional Connectivity (FC) matrix exhibits a strong global signal, making it appear incorrect, as evident in the plot. While the situation seems slightly better for some participants, with correlations reaching -0.4, I believe the underlying problem persists. As this is my first fMRI analysis, I suspect I might be making a fundamental error. Based on the output, I’m considering possibilities such as incorrect motion regression (I would like to clarify that I do not intend to regress out the global signal) or using an atlas resolution that doesn’t match my data. Additionally, I have two basic follow-up questions:
a) Is it correct to assume that the columns X, Y, Z in the confounds file correspond to translations in three directions measured in millimeters?
b) Is it correct to assume that the columns rotX, rotY, rotZ in the confounds file correspond to rotations in three directions measured in radians?
I would be grateful for any advice, particularly from someone familiar with the specifics of the UCLA dataset.
Thank you in advance,
Szymon