Hi there!
I’ve been using QSIPREP to pre-process diffusion MRIs. Here’s the code I used:
singularity run --cleanenv \
-B ${HOME}/EcriPark_Code:/code,${EcriPark}:/data,/scratch/lcorcos/EcriPark_QSIPREP/TESTV9:/out,/home/lcorcos/freesurfer/license.txt,/scratch/lcorcos/Temp_QSIPREP/TMP:/tmp \
--nv ${HOME}/qsiprep-0.18.1.sif /data/ \
/out/ participant --participant_label ${SUB} \
--skip_bids_validation \
--nthreads 24 \
--omp_nthreads 12 \
--fs_license_file /home/lcorcos/freesurfer/license.txt \
--work_dir /tmp/ \
--output_resolution 1 \
--eddy_config /code/eddy_params.json \
--anatomical-template MNI152NLin2009cAsym \
--verbose \
--anat_modality T1w \
--b0_threshold 50 \
--dwi_denoise_window 5 \
--denoise_method dwidenoise \
--unringing_method mrdegibbs \
--distortion_group_merge average \
--bo_motion_corr_to iterative \
--hmc_transform Affine \
--hmc_model eddy \
--skull_strip_template OASIS \
--skull_strip_fixed_seed \
--write_graph
Version:
qsiprep 0.18.1
freesurfer-linux-centos8_x86_64-7.4.1-20230613-7eb8460
singularity 1.1.6-1.el7
Hardware:
Dell PowerEdge C4140 (32 cores) Intel Xeon CPU 5218
380 Go RAM
NVIDIA Tesla V100
The main objective is to use this data to calculate scalar maps (FA, MD, RD, etc…) and to do inter-group comparison. To do this, I imagined putting the data in the same space (MNI) and applying an atlas to it.
The problem is that the data processed by QSIPREP doesn’t seem to be in the MNI space. (I downloaded the ICBM 152 Nonlinear atlases version 2009 here: Templates and atlases — The Princeton Handbook for Reproducible Neuroimaging) and it didn’t fit with the data.
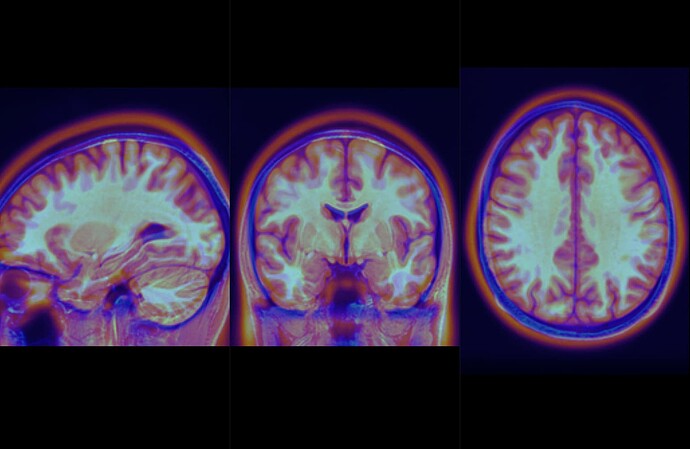
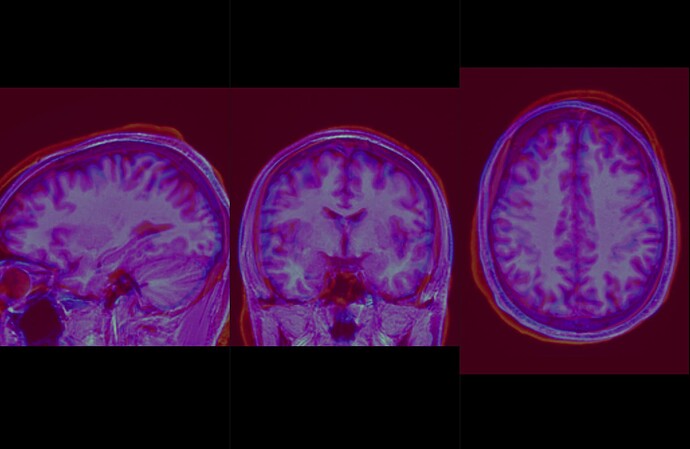
Moreover, each of my subjects didn’t fit between them, and I have the impression that there was no recalibration… (see photos)
Also, here’s the QSIPREP output structure:
├── anat
│ ├── sub-CTR01_desc-aseg_dseg.nii.gz
│ ├── sub-CTR01_desc-brain_mask.nii.gz
│ ├── sub-CTR01_desc-preproc_T1w.nii.gz
│ ├── sub-CTR01_dseg.nii.gz
│ ├── sub-CTR01_from-MNI152NLin2009cAsym_to-T1w_mode-image_xfm.h5
│ ├── sub-CTR01_from-T1wACPC_to-T1wNative_mode-image_xfm.mat
│ ├── sub-CTR01_from-T1wNative_to-T1wACPC_mode-image_xfm.mat
│ └── sub-CTR01_from-T1w_to-MNI152NLin2009cAsym_mode-image_xfm.h5
├── figures
│ ├── sub-CTR01_seg_brainmask.svg
│ ├── sub-CTR01_ses-01_carpetplot.svg
│ ├── sub-CTR01_ses-01_coreg.svg
│ ├── sub-CTR01_ses-01_desc-initial_b0ref.svg
│ ├── sub-CTR01_ses-01_desc-sdc_b0.svg
│ ├── sub-CTR01_ses-01_dir-AP_dwi_denoise_ses_01_dir_AP_dwi_wf_denoising.svg
│ ├── sub-CTR01_ses-01_dir-AP_dwi_denoise_ses_01_dir_AP_dwi_wf_unringing.svg
│ ├── sub-CTR01_ses-01_dir-PA_dwi_denoise_ses_01_dir_PA_dwi_wf_denoising.svg
│ ├── sub-CTR01_ses-01_dir-PA_dwi_denoise_ses_01_dir_PA_dwi_wf_unringing.svg
│ ├── sub-CTR01_ses-01_sampling_scheme.gif
│ └── sub-CTR01_t1_2_mni.svg
└── ses-01
├── anat
│ └── sub-CTR01_ses-01_from-orig_to-T1w_mode-image_xfm.txt
└── dwi
├── sub-CTR01_ses-01_confounds.tsv
├── sub-CTR01_ses-01_desc-ImageQC_dwi.csv
├── sub-CTR01_ses-01_dwiqc.json
├── sub-CTR01_ses-01_space-T1w_desc-brain_mask.nii.gz
├── sub-CTR01_ses-01_space-T1w_desc-eddy_cnr.nii.gz
├── sub-CTR01_ses-01_space-T1w_desc-preproc_dwi.b
├── sub-CTR01_ses-01_space-T1w_desc-preproc_dwi.bval
├── sub-CTR01_ses-01_space-T1w_desc-preproc_dwi.bvec
├── sub-CTR01_ses-01_space-T1w_desc-preproc_dwi.nii.gz
├── sub-CTR01_ses-01_space-T1w_desc-preproc_dwi.txt
└── sub-CTR01_ses-01_space-T1w_dwiref.nii.gz
It differs from what’s marked in the doc (Preprocessing — qsiprep version documentation)
Is this due to out-of-date documentation or have I set the command options incorrectly?
Next, I use MRTRIX’s dwi2tensor and tensor2metric commands to obtain scalar maps. Is this the right choice?
Finally, do I need to run Freesurfer’s recon-all command to obtain a parcelation of the GM and WM? Or should I do a registration of an existing atlas?
Many thanks for your help.