Dear dHCP team,
First thing comes first. Great thank you to your contribution. I was able to download preprocessed fMRI data through the torrent link.
Then, using warping matrix file “~/xfm/sub-XX_ses-XX_from-bold_to-template40wk_mode-image.nii.gz”, which was also downloaded, I successfully warped individual fMRI data into volumetric brain template.
However, my interest is with fMRI data in cortical space, which was downloaded through the link: Atlases from the dHCP project – Brain Development.
I am struggling to find out how I can project volumetric fMRI data into standard cortical fMRI data, without running the whole structural preprocessing pipeline once again. It would be really appreciated if you can let me know.
BTW, when I visualize cortical surfaces of left and right hemisphere (see below), which was downloaded from * Neonatal cortical atlas (270 subjects, Bozek et al. 2018), using Fieldtrip toolbox ft_read_headshape.m, positions of hemisphere was not paired (right hemisphere is obviously higher than left hemisphere), is it because of the compatibility issue? If not, is it intended? 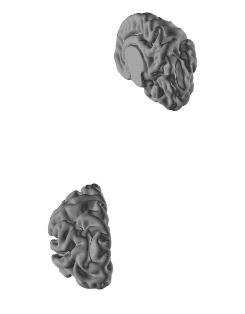
Best,
Jung-Hoon