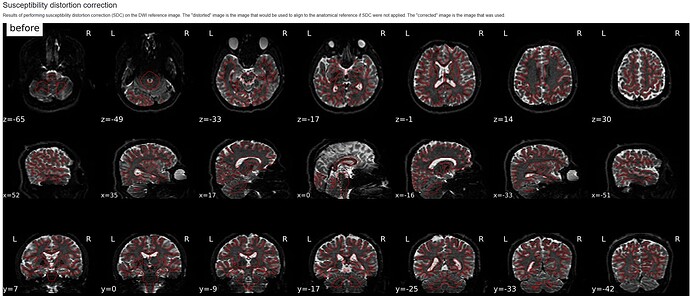
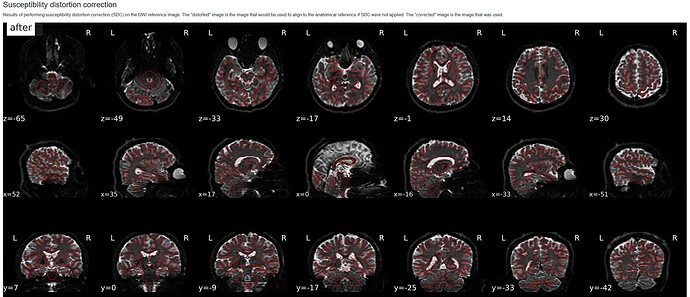
Hello! I am using the qsiprep-1.0.1 Singularity container to preprocess 258-volume DSI data without a fieldmap. However, I found that the parameter --use-syn-sdc did not work.
qsiprep version
1.0.2.dev0+gfc89945.d20250405
Code
--n-cpus 6 --omp-nthreads 6 --mem-mb 120G \
--participant-label sub-0001 \
--fs-license-file /fs_license/license.txt \
--skip-bids-validation \
--subject-anatomical-reference unbiased \
--skip-anat-based-spatial-normalization \
--anat-modality T1w \
--unringing-method mrdegibbs \
--output-resolution 2 \
--hmc-model 3dSHORE \
--use-syn-sdc \
--stop-on-first-crash \
--resource-monitor \
--notrack
Report
Diffusion
Reports for: session 001, run 1.
Summary
Phase-encoding (PE) direction: Anterior-Posterior
Susceptibility distortion correction: None
Coregistration Transform: Rigid
Denoising Method: dwidenoise
Denoising Window: auto
HMC Transform: Affine
HMC Model: 3dSHORE
DWI series resampled to spaces: ACPC
Confounds collected:
Impute slice threshold:
After adding the --force-syn parameter to enforce synthetic fieldmap generation in the QSIPrep workflow, the pipeline encountered an error during the atlas_2_ref step.
code
singularity run --cleanenv \
-B $wd/sub-$subj:/wd \
-B $bids:/bids \
-B $output:/output \
-B $fs_license:/fs_license \
-B $templateflow:$templateflow \
/ibmgpfs/cuizaixu_lab/xulongzhou/apps/singularity/qsiprep-1.0.1.sif \
/bids /output participant -w /wd \
--n-cpus 6 --omp-nthreads 6 --mem-mb 120G \
--participant-label sub-$subj \
--fs-license-file /fs_license/license.txt \
--skip-bids-validation \
--subject-anatomical-reference unbiased \
--skip-anat-based-spatial-normalization \
--anat-modality T1w \
--ignore fieldmaps \
--unringing-method mrdegibbs \
--output-resolution 2 \
--hmc-model 3dSHORE \
--use-syn-sdc error \
--force-syn \
--stop-on-first-crash \
--resource-monitor \
--notrack
rm -rf $wd/sub-$subj
Log
###############################
250527-11:21:45,604 nipype.workflow INFO:
[Node] Executing "atlas_2_ref" <nipype.interfaces.ants.resampling.ApplyTransforms>
250527-11:21:45,611 nipype.workflow INFO:
[Node] Finished "atlas_2_ref", elapsed time 0.000525s.
250527-11:21:45,612 nipype.workflow WARNING:
Storing result file without outputs
250527-11:21:45,614 nipype.workflow WARNING:
[Node] Error on "qsiprep_1_0_wf.sub_0005_ses_001_wf.dwi_preproc_ses_001_run_001_wf.qsiprep_hmcsdc_wf.sdc_wf.syn_sdc_wf.atlas_2_ref" (/wd/qsiprep_1_0_wf/sub_0005_ses_001_wf/dwi_preproc_ses_001_run_001_wf/qsiprep_hmcsdc_wf/sdc_wf/syn_sdc_wf/atlas_2_ref)
250527-11:21:47,420 nipype.workflow ERROR:
Node atlas_2_ref failed to run on host fat17.
250527-11:21:47,424 nipype.workflow ERROR:
Saving crash info to /output/sub-0005/log/20250527-104203_dd84e848-6a53-430b-93cc-0642b1871284/crash-20250527-112147-xulongzhou-atlas_2_ref-2ea41f5d-80ff-4882-bf93-9a0051e51e74.txt
Traceback (most recent call last):
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/pipeline/plugins/multiproc.py", line 66, in run_node
result["result"] = node.run(updatehash=updatehash)
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 525, in run
result = self._run_interface(execute=True)
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 643, in _run_interface
return self._run_command(execute)
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 769, in _run_command
raise NodeExecutionError(msg)
nipype.pipeline.engine.nodes.NodeExecutionError: Exception raised while executing Node atlas_2_ref.
Traceback:
Traceback (most recent call last):
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/interfaces/base/core.py", line 939, in _parse_inputs
arg = self._format_arg(name, spec, value)
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/interfaces/ants/resampling.py", line 495, in _format_arg
return self._get_transform_filenames()
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/interfaces/ants/resampling.py", line 470, in _get_transform_filenames
raise ValueError(
ValueError: ERROR: The invert_transform_flags list must have the same number of entries as the transforms list.
The above exception was the direct cause of the following exception:
Traceback (most recent call last):
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/interfaces/base/core.py", line 739, in _run_interface
runtime.cmdline = self.cmdline
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/interfaces/base/core.py", line 664, in cmdline
allargs = [self._cmd_prefix + self.cmd] + self._parse_inputs()
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/interfaces/base/core.py", line 941, in _parse_inputs
raise ValueError(
ValueError: Error formatting command line argument 'transforms' with value '['/wd/qsiprep_1_0_wf/sub_0005_ses_001_wf/dwi_preproc_ses_001_run_001_wf/qsiprep_hmcsdc_wf/sdc_wf/syn_sdc_wf/ref_2_t1/transform0GenericAffine.mat']'
The above exception was the direct cause of the following exception:
Traceback (most recent call last):
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/interfaces/base/core.py", line 401, in run
runtime = self._run_interface(runtime)
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/interfaces/base/core.py", line 741, in _run_interface
raise RuntimeError(
RuntimeError: Error raised when interpolating the command line
I modified the code to adjust the atlas_2_ref registration step in the QSIPrep pipeline. While this modification allowed the step to complete successfully. I am uncertain whether the normalization process is inherently required for fieldmap-less susceptibility distortion correction (SDC) methods like Syn-SDC.
code
singularity run --cleanenv \
-B $wd/sub-$subj:/wd \
-B $bids:/bids \
-B $output:/output \
-B $fs_license:/fs_license \
-B $templateflow:$templateflow \
/ibmgpfs/cuizaixu_lab/xulongzhou/apps/singularity/qsiprep-1.0.1.sif \
/bids /output participant -w /wd \
--n-cpus 6 --omp-nthreads 6 --mem-mb 120G \
--participant-label sub-$subj \
--fs-license-file /fs_license/license.txt \
--skip-bids-validation \
--subject-anatomical-reference sessionwise \
--anat-modality T1w \
--ignore fieldmaps \
--unringing-method mrdegibbs \
--output-resolution 2 \
--hmc-model 3dSHORE \
--use-syn-sdc error \
--force-syn \
--stop-on-first-crash \
--resource-monitor \
--notrack
Hi @xlzhope,
Thank you for reporting this! This seems like a bug, it would be great if you could post this on our GitHub repo. Would you also mind sharing the solution that you modified? We plan on taking a look at SDC in an upcoming release.
Best,
Steven
OK,I will share this issue in QSIPrep github.
Here is the successful running code.
code
singularity run --cleanenv \
-B $wd/sub-$subj:/wd \
-B $bids:/bids \
-B $output:/output \
-B $fs_license:/fs_license \
-B $templateflow:$templateflow \
/ibmgpfs/cuizaixu_lab/xulongzhou/apps/singularity/qsiprep-1.0.1.sif \
/bids /output participant -w /wd \
--n-cpus 6 --omp-nthreads 6 --mem-mb 120G \
--participant-label sub-$subj \
--fs-license-file /fs_license/license.txt \
--skip-bids-validation \
--subject-anatomical-reference sessionwise \
--anat-modality T1w \
--ignore fieldmaps \
--unringing-method mrdegibbs \
--output-resolution 2 \
--hmc-model 3dSHORE \
--use-syn-sdc error \
--force-syn \
--stop-on-first-crash \
--resource-monitor \
--notrack
Report
* Phase-encoding (PE) direction: Anterior-Posterior
* Susceptibility distortion correction: FLB ("fieldmap-less", SyN-based)
* Coregistration Transform: Rigid
* Denoising Method: dwidenoise
* Denoising Window: auto
* HMC Transform: Affine
* HMC Model: 3dSHORE
* DWI series resampled to spaces: ACPC
* Confounds collected:
* Impute slice threshold:
1 Like