<qsiprep seems to resample the T1s in my dataset really awkwardly. Not sure what’s going on, because there’s no error message. >
singularity run --cleanenv \
/derivatives/BIDS_accession_sub /out participant \
--fs-license-file /opt/freesurfer/license.txt \
--output-resolution 1.2 \
--work-dir
--unringing-method mrdegibbs --denoise-method dwidenoise --nthreads \
--participant_label ${subjectID} \
--separate_all_dwis \
-v -v>
0.19.0
Singularity
BIDS
Summary Available Modalities:
16 Files, 55.41MB FLAIR
1 - Subject T1w
1 - Session dwi
No errors occurred.
Preprocessed T1
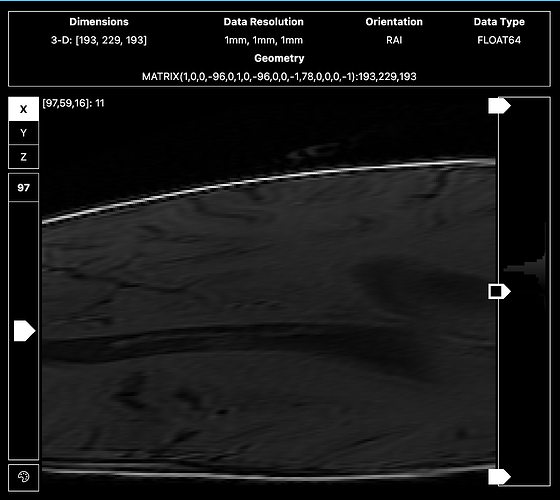
Sharing screenshots of the un-preprocessed T1 below.
jsein
January 16, 2024, 8:40am
3
Hi @suneelbanerjee ,
At first glance, it looks like you are trying to preprocess children images? If it is the case, the normalization to an adult template as it is done by default in qsiprep may fail . You could try with one of the following options:
--anatomical-template and pass a children template as provided in templateflow for instance--infant
It also looks like your T1w might be a localizer or a vnav instead of the 1mm isotropic image most people acquire
Steven
January 16, 2024, 2:06pm
5
Hi @suneelbanerjee and welcome to neurostars!
Adding on to what others have said, you have issues with your command that you should fix before trying again.
You added these flags but you did not specify arguments for them. It should read something like
--work-dir /PATH/TO/WORKDIR --nthreads NUM_THREADS
(making sure that any relevant filesystems are mounted with the -B argument in the Singularity preamble).
suneelbanerjee:
0.19.0
You can try upgrading to 0.20.0
This is only the end of the BIDS validation report. There is usually more above it (errors or warnings if applicable).
Best,
jsein
January 16, 2024, 3:51pm
6
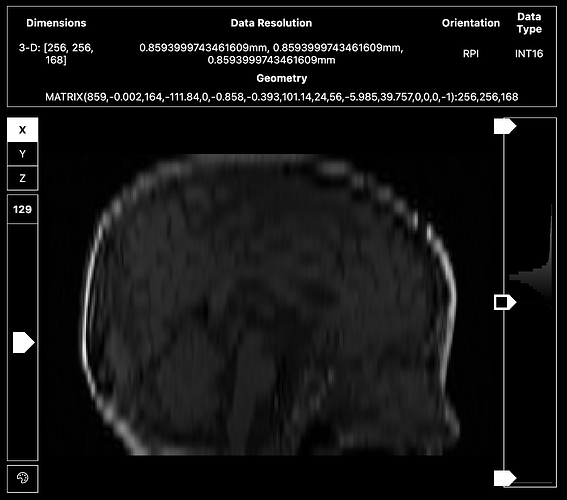
Indeed that is a good catch! It seems like the T1w has indeed very low resolution et and low contrast!
Dear Matt,
Yeah, it’s certainly not the kind of T1 I’d like to be using, but it’s the only one I have available for this subject. It’s a series of axial slices interpolated and upsampled. Do you think that the resampling issue is related to the poor quality of the T1? I have noticed this same issue in a completely different dataset that has high quality T1 and DWI, which is why I think this might be a user error of some kind.
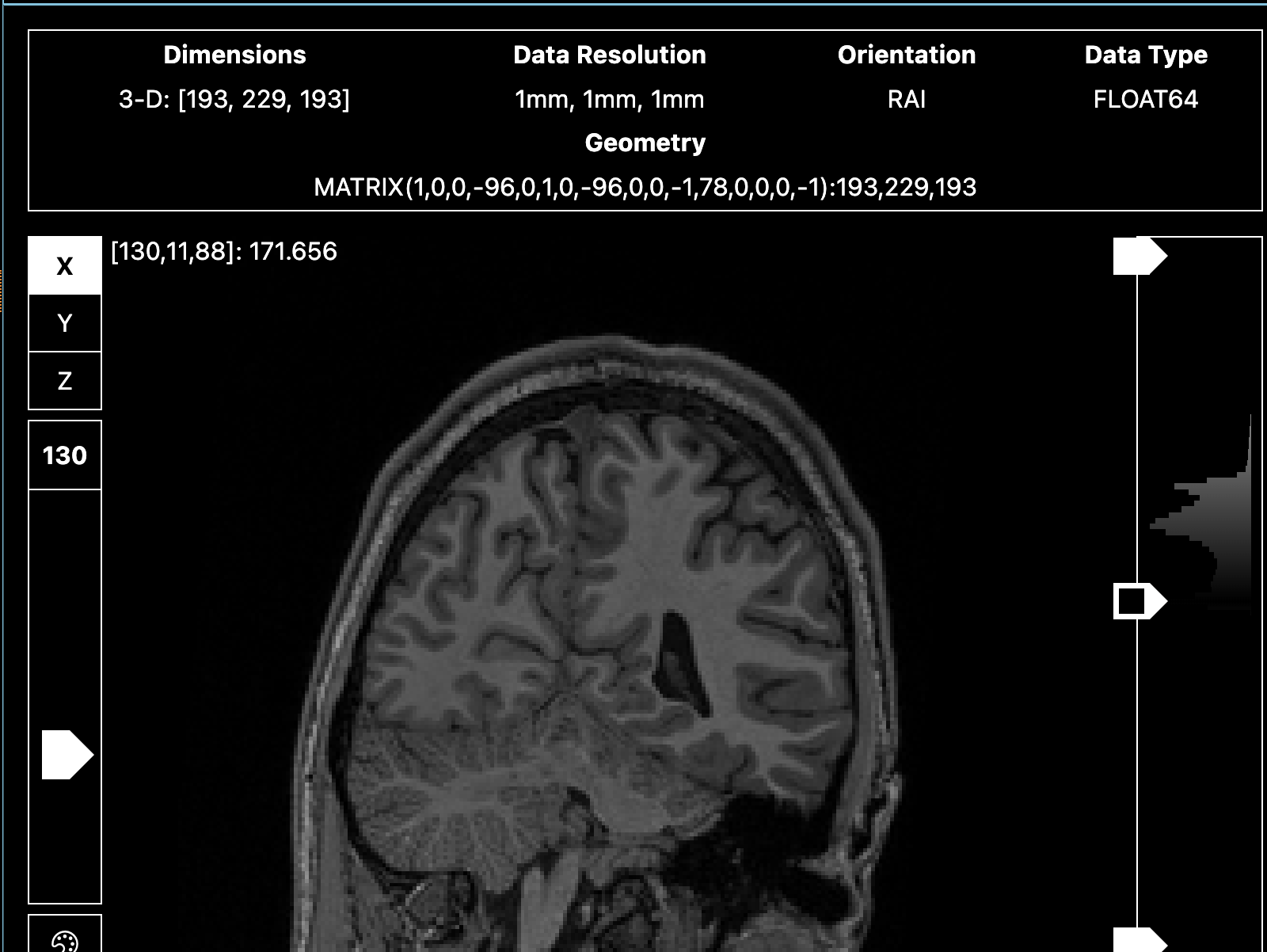
Preprocessed:
Original call here. I specified an anatomical template this time and didn’t use ‘separate all dwis’ because there was only one session.
sudo singularity run --containall --writable-tmpfs -B /media/neel/MOUS/MOUS/MOUS/SynologyDrive/AFQ_working:/data -B /media/neel/dataDrive1/neelbanerjee/qsiprep:/work -B /media/neel/MOUS/MOUS/MOUS/SynologyDrive/AFQ_working/derivatives:/out -B /media/neel/MOUS/MOUS/MOUS/SynologyDrive/DWI_render/license.txt:/opt/freesurfer/license.txt /home/neel/qsiprep-0.19.1.sif /data /out participant --work-dir /work --output-resolution 1.2 --anatomical-template MNI152NLin2009cAsym --fs-license-file /opt/freesurfer/license.txt
Steven
January 19, 2024, 9:31pm
8
Hi @suneelbanerjee ,
Have you tried the —dwi-only (if the poor anatomical outputs are affecting your dwi outputs)?
Best,