Hi everyone!
Summary of what happened:
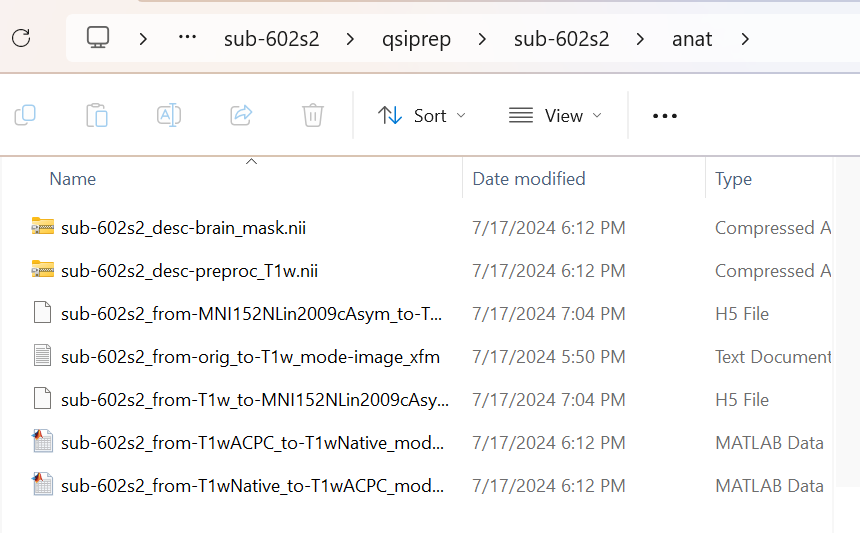
I am running qsiprep on diffusion mri data, using the open-source docker image pennbbl/qsiprep from docker.io/pennbbl/qsiprep. The container runs for 3 hours, creating desc-brain_mask.nii, desc-preproc_T1w.nii and others but is missing the following in the anat folder:
T1w_class-CSF_probtissue.nii.gz
T1w_class-GM_probtissue.nii.gz
T1w_class-WM_probtissue.nii.gz
T1w_dtissue.nii.gz
T1w_preproc.nii.gz
Instead of any of the asym folders, there is just one file that is titled sub___from-MN152NLin2009cAsym_to-T1w-mode-image_xfm.h5 with two accompanying matlab files and a text document with the same T1w-mode-image designation. This mode-image is not present in the documentation’s output, so I think it is an intermediate step that is not fully finishing.
All files that should be present in the dwi folder are present. (According to Preprocessing — qsiprep 0.21.5.dev16+gd9279db documentation)
Command used (and if a helper script was used, a link to the helper script or the command generated):
docker_cmd="docker run --rm \
-v ${BIDS_DIR}:/data \
-v ${OUTPUT_DIR}:/output \
-v ${WORK_DIR}:/work \
-v ${fs_license}:/fs_license.txt \
docker.io/pennbbl/qsiprep"
# Whole command to run, appending specifics to the previous command
cmd="${docker_cmd} /data /output participant\
--skip-bids-validation \
--ignore fieldmaps \
-w /work \
--participant_label sub-${subj_id} \
--output-resolution 2 \
--unringing-method mrdegibbs \
--n_cpus 5 \
--fs-license-file /fs_license.txt"
Environment
docker and VS Code
Data formatted according to a validatable standard?
validated by https://bids-standard.github.io/bids-validator/ and only error is [Code 37] INTENDED_FOR which can be ignored by the argument ignore fieldmaps
Relevant log outputs (up to 20 lines):
QSIPrep failed: Traceback (most recent call last):
........
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 771, in _run_command
raise NodeExecutionError(msg)
nipype.pipeline.engine.nodes.NodeExecutionError: Exception raised while executing Node synthseg.
Cmdline:
mri_synthseg --i /work/qsiprep_wf/single_subject_603s1_wf/anat_preproc_wf/pad_anat_reference_wf/resample_skulled_to_reference/sub-603s1_T1w_lps_trans.nii.gz --threads 1 --post sub-603s1_T1w_lps_trans_post.nii.gz --qc sub-603s1_T1w_lps_trans_qc.csv --o sub-603s1_T1w_lps_trans_aseg.nii.gz
Stderr:
DeprecationWarning: `np.int` is a deprecated alias for the builtin `int`.
Deprecated in NumPy 1.20; for more details and guidance: https://numpy.org/devdocs/release/1.20.0-notes.html#deprecations
DeprecationWarning: `np.float` is a deprecated alias for the builtin `float`.
Deprecated in NumPy 1.20; for more details and guidance: https://numpy.org/devdocs/release/1.20.0-notes.html#deprecations
Killed
Traceback:
.......
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/traits/base_trait_handler.py", line 74, in error
raise TraitError(
traits.trait_errors.TraitError: The 'out_post' trait of a _SynthSegOutputSpec instance must be a pathlike object or string representing an existing file, but a value of '/work/qsiprep_wf/single_subject_603s1_wf/anat_preproc_wf/synthseg_anat_wf/synthseg/sub-603s1_T1w_lps_trans_post.nii.gz' <class 'str'> was specified.
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/interfaces/base/core.py", line 460, in aggregate_outputs
raise FileNotFoundError(msg)
FileNotFoundError: No such file or directory '/work/qsiprep_wf/single_subject_603s1_wf/anat_preproc_wf/synthseg_anat_wf/synthseg/sub-603s1_T1w_lps_trans_post.nii.gz' for output 'out_post' of a SynthSeg interface
Screenshots / relevant information:
I tried running my next two scripts which 1. puts the output of qsiprep into a bids format and 2. runs reconstruction. The bids formatting file runs fine, but the reconstruction errors out.
I wonder if anyone knows of a better qsiprep docker container to run, one with python files with updated syntax. Or, if there is a way to access the scripts that the docker container pulls. I have attached a screenshot of the anat folder in case it is helpful to look at.
Any help is much appreciated!
Thank you so much,
Angelica