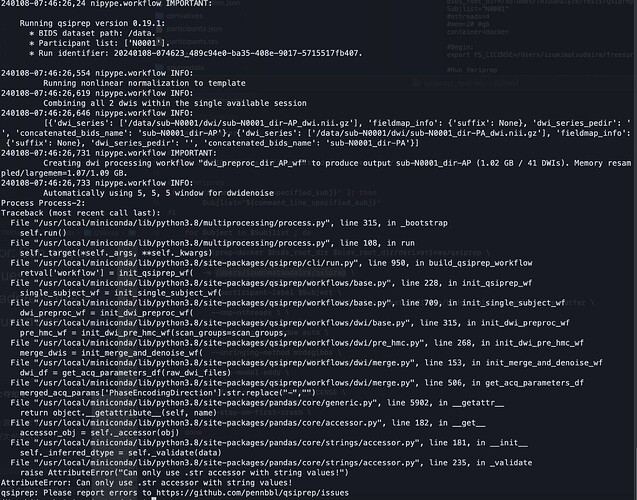
Dear @Steven
Thank you for your reply.
Our DWI JSON file is below.
{
"Modality": "MR",
"MagneticFieldStrength": 3,
"ImagingFrequency": 127.765,
"Manufacturer": "Philips",
"ManufacturersModelName": "Achieva dStream",
"InstitutionName": "TohokuUniv.BrainDynamicStudy",
"InstitutionalDepartmentName": "Rediology",
"DeviceSerialNumber": "17556",
"StationName": "mrhostx",
"BodyPartExamined": "BRAIN",
"PatientPosition": "HFS",
"SoftwareVersions": "5.7.1\\5.7.1.3",
"MRAcquisitionType": "2D",
"SeriesDescription": "DTI Jones30_a",
"ProtocolName": "DTI Jones30_a",
"ScanningSequence": "SE",
"SequenceVariant": "SK",
"ScanOptions": "FS",
"PulseSequenceName": "DwiSE",
"ImageType": [
"ORIGINAL",
"PRIMARY",
"M",
"SE",
"M",
"SE"
],
"SeriesNumber": 701,
"AcquisitionTime": "18:28:53.210000",
"AcquisitionNumber": 7,
"PhilipsRWVSlope": 0.82442,
"PhilipsRWVIntercept": 0,
"PhilipsRescaleSlope": 0.82442,
"PhilipsRescaleIntercept": 0,
"PhilipsScaleSlope": 0.0267161,
"UsePhilipsFloatNotDisplayScaling": 1,
"SliceThickness": 2,
"SpacingBetweenSlices": 2,
"SAR": 0.0606958,
"EchoTime": 0.08,
"RepetitionTime": 10,
"MTState": false,
"FlipAngle": 90,
"CoilString": "MULTI COIL",
"PercentPhaseFOV": 100,
"PercentSampling": 100,
"EchoTrainLength": 55,
"PartialFourierDirection": "PHASE",
"PartialFourierEnabled": "YES",
"PhaseEncodingStepsNoPartialFourier": 110,
"AcquisitionMatrixPE": 110,
"ReconMatrixPE": 112,
"ParallelReductionFactorInPlane": 2,
"ParallelAcquisitionTechnique": "SENSE",
"WaterFatShift": 13.7088,
"EstimatedEffectiveEchoSpacing": 0.00027923,
"EstimatedTotalReadoutTime": 0.0309945,
"AcquisitionDuration": 330,
"PixelBandwidth": 2570,
"PhaseEncodingAxis": "j",
"ImageOrientationPatientDICOM": [
0.997373,
0.0279108,
0.0668486,
-0.0293151,
0.999368,
0.0201182
],
"InPlanePhaseEncodingDirectionDICOM": "COL",
"ConversionSoftware": "dcm2niix",
"ConversionSoftwareVersion": "v1.0.20220720",
"Dcm2bidsVersion": "2.1.7"
}