Summary of what happened:
HI, I am new to QSIprep and Docker. I am running qsiprep with docker container for a single HCP subject. But I get EOS: No space left on device. Which would be the best practice to solve the problem in this case?
Command used
docker run -ti --rm -v $HOME/100307:/data -v $HOME/dti-preproc:/out -v \
licence.txt:/opt/freesurfer/license.txt pennlinc/qsiprep:latest /data /out \
participant --fs-license-file licence.txt --output-resolution 1.2 \
--nthreads 64 --omp-nthreads 2 \
--anat-modality T1w --denoise-after-combining \
--distortion-group-merge average --hmc-model eddy
Version:
qsiprep latest version (1.0.0rc2)
Environment (Docker, Singularity / Apptainer, custom installation):
Docker
Relevant log outputs (up to 20 lines):
241130-04:45:04,744 nipype.workflow WARNING:
[Node] Error on "qsiprep_1_0_wf.sub_100307_wf.dwi_preproc_acq_dir97lr_wf.pre_hmc_wf.merge_and_denoise_wf.merged_denoise.denoiser" (/ssd1/work/qsiprep_1_0_wf/sub_100307_wf/dwi_preproc_acq_dir97lr_wf/pre_hmc_wf/merge_and_denoise_wf/merged_denoise/denoiser)
exception calling callback for <Future at 0x7f2d02447430 state=finished raised FileNotFoundError>
concurrent.futures.process._RemoteTraceback:
"""
Traceback (most recent call last):
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/pipeline/plugins/multiproc.py", line 66, in run_node
result["result"] = node.run(updatehash=updatehash)
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 525, in run
result = self._run_interface(execute=True)
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 643, in _run_interface
return self._run_command(execute)
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 743, in _run_command
_save_resultfile(
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/pipeline/engine/utils.py", line 234, in save_resultfile
savepkl(resultsfile, result)
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/utils/filemanip.py", line 697, in savepkl
with pkl_open(tmpfile, "wb") as pkl_file:
File "/opt/conda/envs/qsiprep/lib/python3.10/gzip.py", line 344, in close
myfileobj.close()
OSError: [Errno 28] No space left on device
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File "/opt/conda/envs/qsiprep/lib/python3.10/concurrent/futures/process.py", line 246, in _process_worker
r = call_item.fn(*call_item.args, **call_item.kwargs)
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/pipeline/plugins/multiproc.py", line 69, in run_node
result["result"] = node.result
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 221, in result
return _load_resultfile(
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/pipeline/engine/utils.py", line 290, in load_resultfile
raise FileNotFoundError(results_file)
FileNotFoundError: /ssd1/work/qsiprep_1_0_wf/sub_100307_wf/dwi_preproc_acq_dir97lr_wf/pre_hmc_wf/merge_and_denoise_wf/merged_denoise/denoiser/result_denoiser.pklz
"""
The above exception was the direct cause of the following exception:
Traceback (most recent call last):
File "/opt/conda/envs/qsiprep/lib/python3.10/concurrent/futures/_base.py", line 342, in _invoke_callbacks
callback(self)
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/pipeline/plugins/multiproc.py", line 158, in _async_callback
result = args.result()
File "/opt/conda/envs/qsiprep/lib/python3.10/concurrent/futures/_base.py", line 451, in result
return self.__get_result()
File "/opt/conda/envs/qsiprep/lib/python3.10/concurrent/futures/_base.py", line 403, in __get_result
raise self._exception
FileNotFoundError: /ssd1/work/qsiprep_1_0_wf/sub_100307_wf/dwi_preproc_acq_dir97lr_wf/pre_hmc_wf/merge_and_denoise_wf/merged_denoise/denoiser/result_denoiser.pklz
Other relevant information
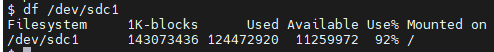
This is the disk that gets filled up to 100% during the process.