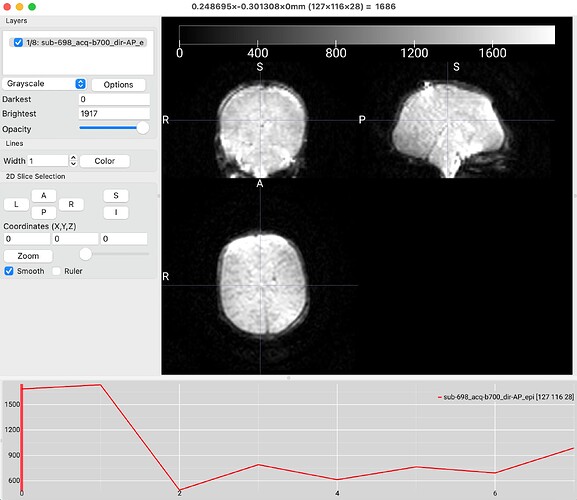
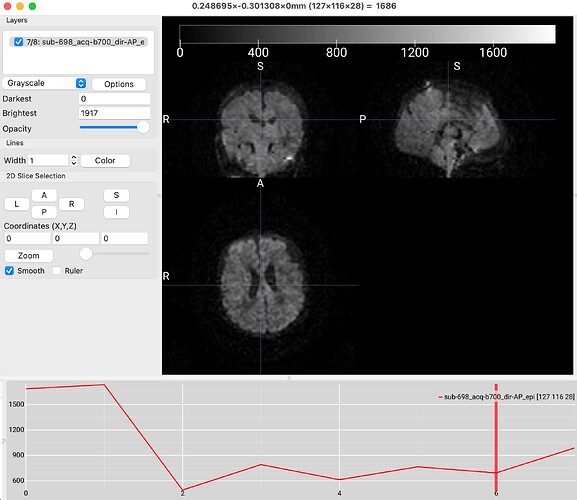
Summary of what happened:
From the citation boilerplate
“A total of 2 distortion groups were included in the data data. Distortion group ‘0 1 0 0.063080’ was represented by image 1 from sub-698_acq-b700_dir-PA_dwi.nii.gz. Distortion group ‘0 -1 0 0.063080’ was represented by image 7 from sub-698_acq-b700_dir-AP_epi.nii.gz.”
I understand from the documentation that the algorithm performs a calculation similar to the developmental HCP pipelines based on pairwise spatial correlations, but I was wondering whether it was possible to tell qsiprep which images to use for TOPUP? For example, I would like to use the image 1 (first image attached) to TOPUP instead of image 7 (second image attached), would that be possible?
When I examined the confounds tsv file (in /qsiprep/<subID>/dwi/), I also could not see a “selected_for_topup” column, so I was wondering if there was another confounds tsv file I was not locating.
(The column headers I have are: framewise_displacement, trans_x, trans_y, trans_z, rot_x, rot_y, rot_z, eddy_stdevs, original_file, grad_x, grad_y, grad_z, bval, image_mean, series_b0_mean, series_b0_correction, DWIDenoise_pre, DWIDenoise_post, DWIDenoise_change)
Command used (and if a helper script was used, a link to the helper script or the command generated):
export APPTAINERENV_FS_LICENSE=$GROUP_HOME/freesurferlicense.txt
singularity run -e --containall --writable-tmpfs --cleanenv \
/home/groups/jyeatman/software/singularity_images/qsiprep-0.20.0.sif \
/scratch/groups/jyeatman/howard/DBP3/bidsfull \
/scratch/groups/jyeatman/howard/DBP3/qsiprepfull participant --participant-label sub-$SLURM_ARRAY_TASK_ID \
-w /scratch/groups/jyeatman/howard/DBP3/qsiprepfull-work \
--nthreads 10 \
--omp-nthreads 8 \
--output-resolution 1.2 \
--infant \
--pepolar-method TOPUP+DRBUDDI
Version:
qsiprep-0.20.0.sif
Environment (Docker, Singularity / Apptainer, custom installation):
Singularity
Data formatted according to a validatable standard? Please provide the output of the validator:
-
Relevant log outputs (up to 20 lines):
-