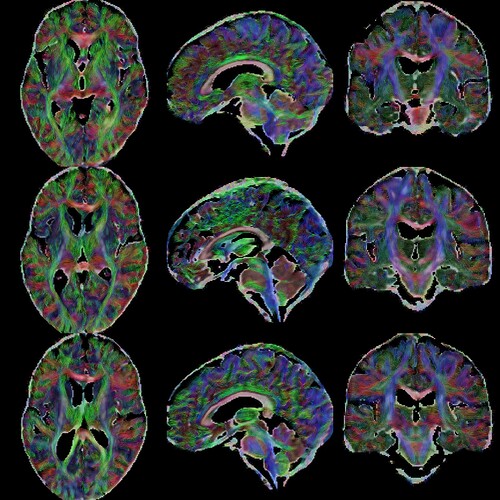
Summary of what happened:
I ran preprocessing with QSIPrep 0.21.4 and NODDI reconstruction with QSIPrep 0.19.1. The figures in the NODDI outputs appear to contain lots of non-brain tissue (dura; see attached figure). I am looking for ways to improve the quality of the NODDI outputs.
I think the source of the issue is the mask that antsBrainExtraction estimates during preprocessing. This mask looks imprecise, and it has noticeable differences (it is bigger and rounder) when compared to a mask created from the subject’s T1 using HD-BET.
Is there any way within QSIPrep to improve the quality of the mask generated during preprocessing?
If not, I have an idea about how to work around this issue, but I an unsure if it is valid. This involves generating a mask from the T1 using HD-BET; using the inverse of ANTS transformations generated during preprocessing to put the new mask into diffusion space; replacing the old DWI mask (the one generated by qsiprep) with the new mask; and rerunning QSIRecon. Would this work (conceptually)?
Thank you!
Command used (and if a helper script was used, a link to the helper script or the command generated):
Preprocessing command:
docker run --rm -it \
-v /home/john/Documents/license.txt:/opt/freesurfer/license.txt:ro \
-v /home/john/Documents/ADNI_Multishell_SCD_2024/ADNI_bids/bidsconvertr/bids:/data:ro \
-v /home/john/Documents/ADNI_Multishell_SCD_2024/ADNI_bids/bidsconvertr/derivatives:/out \
-v /home/john/Documents/ADNI_Multishell_SCD_2024/ADNI_bids/bidsconvertr/derivatives/work:/scratch \
pennbbl/qsiprep:0.21.4 /data /out participant \
--participant_label $PARTICIPANT_ID \
--output-resolution 1 \
--nthreads 24 \
--unringing-method mrdegibbs \
--force-syn \
-w /scratch
Recon command:
docker run --rm \
-v ~/Documents/license.txt:/opt/freesurfer/license.txt:ro \
-v ~/Documents/ADNI_Multishell_SCD_2024/ADNI_bids/bidsconvertr/bids:/data:ro \
-v ~/Documents/ADNI_Multishell_SCD_2024/ADNI_bids/bidsconvertr/derivatives:/qsiprep:ro \
-v ~/Documents/Docker_Commands/amico_noddi_cortex.json:/sngl/spec/spec.json:ro \
-v ~/Documents/ADNI_Multishell_SCD_2024/ADNI_bids/bidsconvertr/derivatives:/out \
-v ~/Documents/ADNI_Multishell_SCD_2024/ADNI_bids/bidsconvertr/derivatives/work:/scratch \
pennbbl/qsiprep:0.19.1 /data /out participant \
--recon-only \
--recon-input /qsiprep \
--recon-spec /sngl/spec/spec.json \
--participant_label $PARTICIPANT_ID \
--output-resolution 1 \
--nthreads 24 \
-w /scratch
Version:
QSIPrep 0.21.4 for preprocessing
QSIPrep 0.19.1 for reconstruction
Environment (Docker, Singularity / Apptainer, custom installation):
Docker
Data formatted according to a validatable standard? Please provide the output of the validator:
Using bids-validator v1.14.14, I get 0 errors and 2 types of warning:
1: [WARN] Not all subjects contain the same files. Each subject should contain the same number of files with the same naming unless some files are known to be missing. (code: 38 - INCONSISTENT_SUBJECTS)
2: [WARN] Not all subjects/sessions/runs have the same scanning parameters.