Summary of what happened:
I am trying to use the QSIprep Preprocessing pipeline but the execution quickly crashes
Command used (and if a helper script was used, a link to the helper script or the command generated):
I used command:
qsiprep /media/zhangming/E88686698686708/test/wx/test_out /media/zhangming/E88686698686708/test/wx/test_out/derivatives participant participant-label 001 --skip_bids_validation --output-resolution 2 --fs-license-flie /media/zhangming/E88686698686708/test/wx/test_out/license
Version:
QSIprep Version:0.16.1
Environment (Docker, Singularity, custom installation):
Environment: Docker
Data formatted according to a validatable standard? Please provide the output of the validator:
Data formatted according to BIDS standard
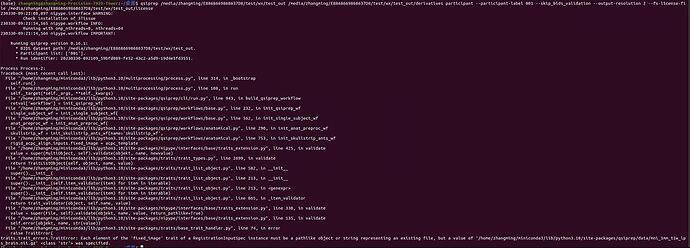
Relevant log outputs (up to 20 lines):
230330-12:18:23,720 nipype.interface WARNING:
Check installation of 3Tissue
230330-12:18:29,187 nipype.workflow INFO:
Running with omp_nthreads=8, nthreads=64
230330-12:18:29,188 nipype.workflow IMPORTANT:
Running qsiprep version 0.16.1:
* BIDS dataset path: /media/zhangming/E8868669868637D8/test/wx/test_out.
* Participant list: ['001'].
* Run identifier: 20230330-121824_8be945af-c9c1-45c3-acbd-5a8495e73d80.
Process Process-2:
Traceback (most recent call last):
File "/home/zhangming/miniconda3/lib/python3.10/multiprocessing/process.py", line 314, in _bootstrap
self.run()
File "/home/zhangming/miniconda3/lib/python3.10/multiprocessing/process.py", line 108, in run
self._target(*self._args, **self._kwargs)
File "/home/zhangming/miniconda3/lib/python3.10/site-packages/qsiprep/cli/run.py", line 943, in build_qsiprep_workflow
retval['workflow'] = init_qsiprep_wf(
File "/home/zhangming/miniconda3/lib/python3.10/site-packages/qsiprep/workflows/base.py", line 232, in init_qsiprep_wf
single_subject_wf = init_single_subject_wf(
File "/home/zhangming/miniconda3/lib/python3.10/site-packages/qsiprep/workflows/base.py", line 562, in init_single_subject_wf
anat_preproc_wf = init_anat_preproc_wf(
File "/home/zhangming/miniconda3/lib/python3.10/site-packages/qsiprep/workflows/anatomical.py", line 290, in init_anat_preproc_wf
skullstrip_wf = init_skullstrip_ants_wf(name='skullstrip_wf',
File "/home/zhangming/miniconda3/lib/python3.10/site-packages/qsiprep/workflows/anatomical.py", line 753, in init_skullstrip_ants_wf
rigid_acpc_align.inputs.fixed_image = acpc_template
File "/home/zhangming/miniconda3/lib/python3.10/site-packages/nipype/interfaces/base/traits_extension.py", line 425, in validate
value = super(MultiObject, self).validate(objekt, name, newvalue)
File "/home/zhangming/miniconda3/lib/python3.10/site-packages/traits/trait_types.py", line 2699, in validate
return TraitListObject(self, object, name, value)
File "/home/zhangming/miniconda3/lib/python3.10/site-packages/traits/trait_list_object.py", line 582, in __init__
super().__init__(
File "/home/zhangming/miniconda3/lib/python3.10/site-packages/traits/trait_list_object.py", line 213, in __init__
super().__init__(self.item_validator(item) for item in iterable)
File "/home/zhangming/miniconda3/lib/python3.10/site-packages/traits/trait_list_object.py", line 213, in <genexpr>
super().__init__(self.item_validator(item) for item in iterable)
File "/home/zhangming/miniconda3/lib/python3.10/site-packages/traits/trait_list_object.py", line 865, in _item_validator
return trait_validator(object, self.name, value)
File "/home/zhangming/miniconda3/lib/python3.10/site-packages/nipype/interfaces/base/traits_extension.py", line 330, in validate
value = super(File, self).validate(objekt, name, value, return_pathlike=True)
File "/home/zhangming/miniconda3/lib/python3.10/site-packages/nipype/interfaces/base/traits_extension.py", line 135, in validate
self.error(objekt, name, str(value))
File "/home/zhangming/miniconda3/lib/python3.10/site-packages/traits/base_trait_handler.py", line 74, in error
raise TraitError(
traits.trait_errors.TraitError: Each element of the 'fixed_image' trait of a RegistrationInputSpec instance must be a pathlike object or string representing an existing file, but a value of '/home/zhangming/miniconda3/lib/python3.10/site-packages/qsiprep/data/mni_1mm_t1w_lps_brain.nii.gz' <class 'str'> was specified.
Screenshots / relevant information:
QSIprep installation following:
docker pull PennLINC/qsiprep
pip install --user --upgrade qsiprep-container.
Can somebody point to what I am doing wrong?
Thank you!