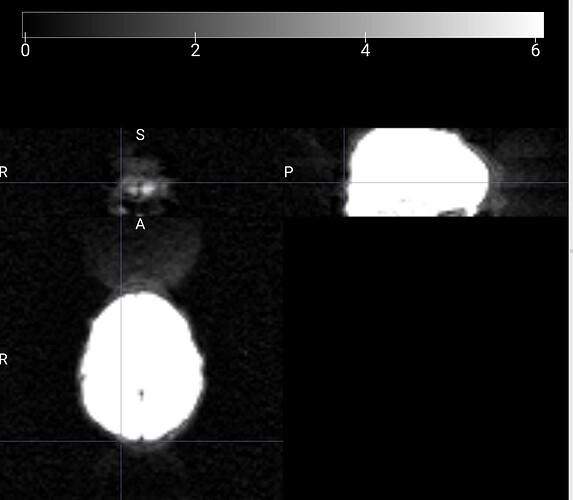
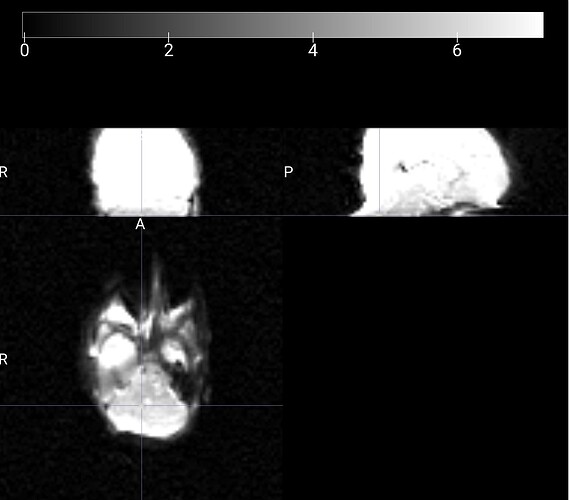
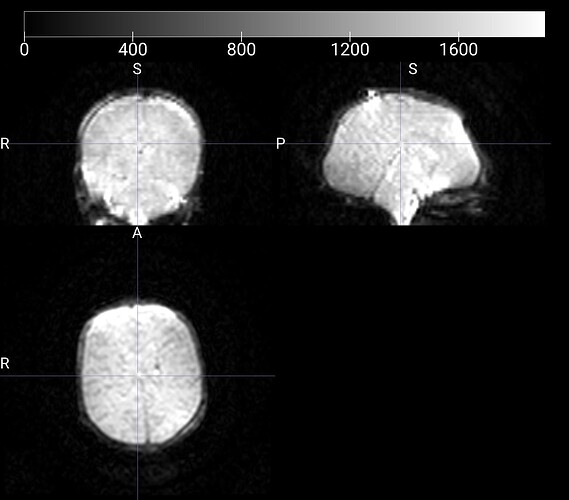
Summary of what happened:
I am trying to run qsiprep 0.20.0 on a single subject with one T1w image and one dwi image. I am having difficulty understanding the error flagged in the log during the denoising step, and was hoping to get some help with it.
“ValueError: zero-size array to reduction operation minimum which has no identity”
(I’ve tagged both nipype and qsiprep because I can’t tell which is the cause)
Command used (and if a helper script was used, a link to the helper script or the command generated):
export APPTAINERENV_FS_LICENSE=$GROUP_HOME/freesurferlicense.txt
singularity run -e --containall --writable-tmpfs --cleanenv \
/home/groups/jyeatman/software/singularity_images/qsiprep-0.20.0.sif \
/scratch/groups/jyeatman/howard/DBP2/bids \
/scratch/groups/jyeatman/howard/DBP2/qsiprep participant --participant-label sub-$SLURM_ARRAY_TASK_ID \
-w /scratch/groups/jyeatman/howard/DBP2/qsiprep-work \
--nthreads 10 \
--omp-nthreads 8 \
--output-resolution 1.2 \
--infant \
--pepolar-method TOPUP+DRBUDDI
Version:
singularity build from docker://pennbbl/qsiprep:0.20.0 on HPC cluster
Environment (Docker, Singularity / Apptainer, custom installation):
Apptainer
Data formatted according to a validatable standard? Please provide the output of the validator:
Data BIDSified with ezBIDS and validated on https://bids-standard.github.io/bids-validator/
Summary
10 Files, 85.75MB
1 - Subject
1 - Session
Available Tasks
Available Modalities
MRI
We found 1 Warning in your dataset.
view 1 warning
Warning 1: [Code 113] NO_AUTHORS
Click here for more information about this issue
The Authors field of dataset_description.json should contain an array of fields - with one author per field. This was triggered because there are no authors, which will make DOI registration from dataset metadata impossible.
Relevant log outputs (up to 20 lines):
Node: qsiprep_wf.single_subject_21337_wf.dwi_preproc_acq_b700_dir_PA_wf.pre_hmc_wf.merge_and_denoise_wf.dwi_denoise_acq_b700_dir_PA_dwi_wf.denoiser
Working directory: /scratch/groups/jyeatman/howard/DBP2/qsiprep-work/qsiprep_wf/single_subject_21337_wf/dwi_preproc_acq_b700_dir_PA_wf/pre_hmc_wf/merge_and_denoise_wf/dwi_denoise_acq_b700_dir_PA_dwi_wf/denoiser
Node inputs:
args = <undefined>
bval_scale = <undefined>
environ = {}
extent = (5, 5, 5)
grad_file = <undefined>
grad_fsl = <undefined>
in_bval = <undefined>
in_bvec = <undefined>
in_file = <undefined>
mask = <undefined>
nmse_text = <undefined>
noise_image = <undefined>
nthreads = 1
out_bval = <undefined>
out_bvec = <undefined>
out_file = <undefined>
out_report = dwidenoise_report.svg
Traceback (most recent call last):
File "/usr/local/miniconda/lib/python3.10/site-packages/nipype/pipeline/plugins/multiproc.py", line 67, in run_node
result["result"] = node.run(updatehash=updatehash)
File "/usr/local/miniconda/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 527, in run
result = self._run_interface(execute=True)
File "/usr/local/miniconda/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 645, in _run_interface
return self._run_command(execute)
File "/usr/local/miniconda/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 771, in _run_command
raise NodeExecutionError(msg)
nipype.pipeline.engine.nodes.NodeExecutionError: Exception raised while executing Node denoiser.
[…]
dwidenoise: [ 50%] compressing image "sub-21337_acq-b700_dir-PA_dwi_LAS_noise.nii.gz"...e[0Ke[?7he[?7l
dwidenoise: [100%] compressing image "sub-21337_acq-b700_dir-PA_dwi_LAS_noise.nii.gz"...e[0Ke[?7h
dwidenoise: [100%] compressing image "sub-21337_acq-b700_dir-PA_dwi_LAS_noise.nii.gz"e[0K
Traceback:
Traceback (most recent call last):
File "/usr/local/miniconda/lib/python3.10/site-packages/nipype/interfaces/base/core.py", line 398, in run
runtime = self._post_run_hook(runtime)
File "/usr/local/miniconda/lib/python3.10/site-packages/nipype/interfaces/mixins/reporting.py", line 50, in _post_run_hook
self._generate_report()
File "/usr/local/miniconda/lib/python3.10/site-packages/qsiprep/interfaces/denoise.py", line 90, in _generate_report
cuts = cuts_from_bbox(contour_nii or mask_nii, cuts=self._n_cuts)
File "/usr/local/miniconda/lib/python3.10/site-packages/qsiprep/niworkflows/viz/utils.py", line 205, in cuts_from_bbox
start_coords = B.min(0)
File "/usr/local/miniconda/lib/python3.10/site-packages/numpy/core/_methods.py", line 44, in _amin
return umr_minimum(a, axis, None, out, keepdims, initial, where)
ValueError: zero-size array to reduction operation minimum which has no identity