Hi all, apologies for spamming this space with questions. I am new to using rapidtide and am trying to get a better understanding of what is happening under the hood. The question I have here is related to the —searchrange flag/its theory of operation. I went through the rapidtide documentation on what this flag does/how it works but had some lingering questions.
Data and base command: I am extracting whole brain lags for multi-band rsfMRI data (TR=0.46s). My cohort includes young (20s) and older adults (60+). As a starting point, I processed 10 subjects across this age range (their brains looked fairly healthy - no obvious pathology), using this base command:
CMD="apptainer exec -B /path_to/fmriprep/sub-${subjID}/ses-01:/data_in \
-B /path_to/rapidtide_test_search_range_neg20_to_40/sub-${subjID}:/data_out \
/path_to/Software/sif/rapidtide.sif rapidtide \
/data_in/func/sub-${subjID}_ses-01_task-rest_desc-preproc_bold.nii.gz \
/data_out/sub-${subjID}
--numnull 10000 \
--filterband lfo \
--searchrange -20 40 \
--passes 3 \
--despecklepasses 4 \
--acfix \
--nofitfilt \
--brainmask /data_in/func/sub-${subjID}_ses-01_task-rest_desc-brain_mask.nii.gz \
--graymattermask /data_in/fMRIPrep_parc_bold_space/sub-${subjID}_ses-01_task-rest_space-bold_desc-GM_probseg_bin.nii.gz \
--whitemattermask /data_in/fMRIPrep_parc_bold_space/sub-${subjID}_ses-01_task-rest_space-bold_desc-WM_probseg_bin.nii.gz \
--mklthreads 1 \
--nprocs 1 \
--numskip 0 \
--outputlevel max"
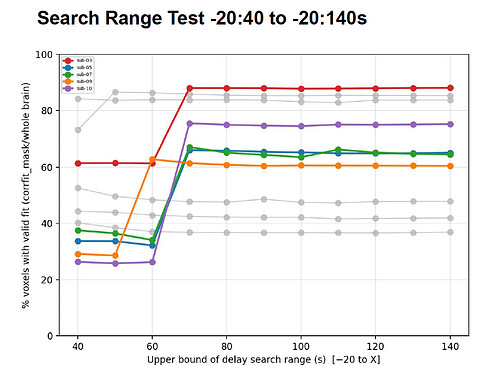
What I tested: I re-ran this command, expanding only the upper limit of the search range, from -20:40 all the way to -20:140s. I then plotted the % of voxels that were determined to have a “valid fit” (corrfit_mask/brainmask) across each search-range window.
Result:
- I noticed that as I increased the upper bound, some subjects had this sharp jump in % of voxels with a valid fit. Note: These subjects were of various age groups (some young, some old).
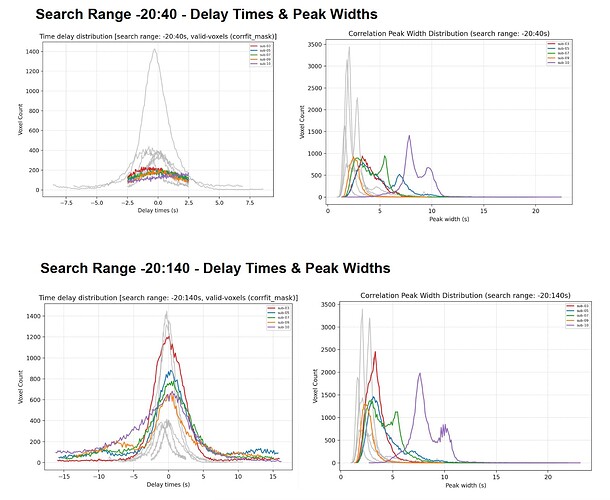
- When plotting a histogram of delay times (-20:40s) vs (-20:140s) for the few that jumped, I noticed that while the % fit increased, the distribution of delay times did not really increase. What was odd was that the window of -20:40 picked up delays of max +/- 7 sec, but a window of -20:140s picked up delays of +/- 15 sec. This was weird, and I wondered if those additionally detected delays were spurious fits.
- I plotted the peak widths for those few subjects, and noticed they were a little broad. The similarity metrics (per
runqualitycheckwere also centered around 0.3-0.6).
Questions:
- Is the pattern I’m seeing spurious? Would it be advisable to keep the search range as wide as possible just to get more fits - or is that hacky?
- In the
runqualitychecksection of the documentation, would it be possible to provide some notes on how to quality check the lag maps? And what you should definitely flag as a fail?
(Sidenote:
-
I also checked the probe regressor time course - I noticed the documentation said that folks can take a solid 60-70s to settle into the scanner, and that might be why setting a long search range is picking up more fits? I tried toggling with
—simcalcrange130 -1 (60s onwards) at the -20:40s search window, but this did not really improve the % of fits so I dropped that. I also tried a few runs with the motion file (6 confounds) supplied. This did not really improve the % of fits either. -
I am currently running some subjects with obvious pathology (CSVD-like/stroke lesions), to see if a wide window picks up longer delays as literature has previuosly shown, maybe this would justify a large window, but I’m not sure).
Apologies for the wall of text. I would greatly appreciate any thoughts on this, thanks!