Hello
I have applied MVPA using NiLearn to my data over the weekend and now I have some questions. Would greatly appreciate some feedback/further information/resources on these.
-
I have more features than examples. I think this must be common in MVPA? What are some of the issues/considerations with this? Which algorithms are appropriate other than SVC (assuming SVC is)?
-
What do we need to do about correlated features in MVPA? Voxel signals violate the independence assumption. Maybe if we use some data reduction technique this might also alleviate #1? But then it might render things less interpretable.
-
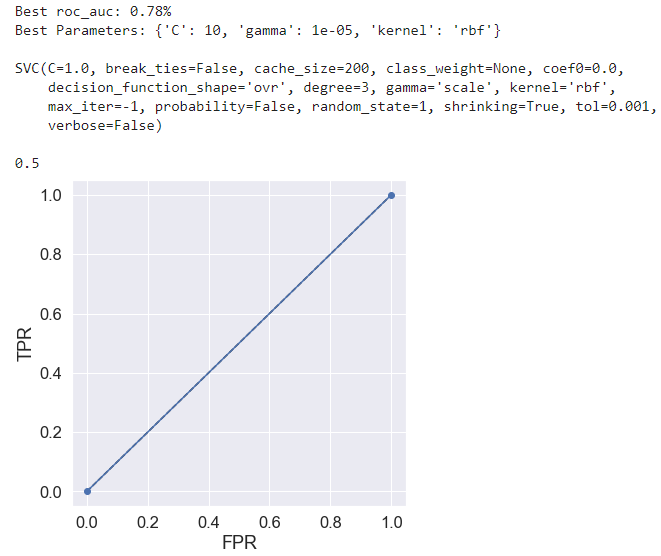
Is it ok to use less interpretable algorithms for classification, sacrificing ability to visualise the results? I found that a linear SVC kernel performed at ~chance level but changing this to RBF and tuning with grid search gave me almost 70% accuracy.
Thanks!