Hi all,
I’ve been running fMRIPREP with fieldmap data to include susceptibility distortion correction (SDC) and have a few questions about the output of susceptibility distortion correction.
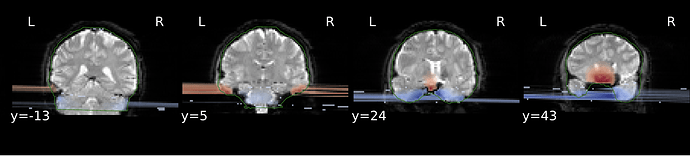
I’m not sure how the SDC output is supposed to look like as this is my first time running fMRIPREP with field map data. Below is what I’ve got from one participant:
It seems that these blue and red lines are causing more distortion (like dragging the bottom parts of brain to the left) in EPI data which field map data were applied to. This looks a bit problematic as the distorted EPI images are not aligned well with ANAT.
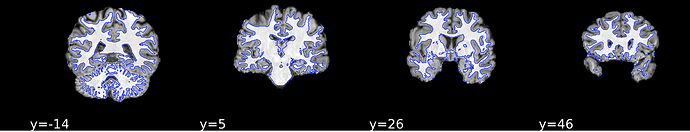
Here’s the ANAT images from the same participant’s html report:
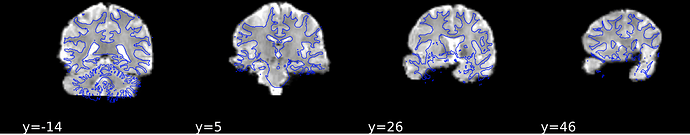
And EPI images:
So I was wondering if this dragging issue is normal after SDC?
I’d really appreciate your help or comments. Thank you.
Thank you so much for your reply.
I now understand that the rays in the SDC output are fine.
But I’m still not sure why SDC affected the alignment between EPI and ANAT (especially the bottom parts of the brain).
Is there anything I can do to fix the alignment? Or do you think the alignment looks fine?
Thank you in advance!
Oh right, I didn’t scroll enough to check that in detail. I think that the warp is actually being applied in the opposite direction it should. There’s a possibility that fMRIPrep is taking it wrong, but most likely your metadata is not correct (the PhaseEncodingDirection in particular).
That makes sense! I will double-check my metadata. Thanks so much!
Just one last question, could you let me know how I can check whether the direction is opposite or not?
I used HeuDiConv with the ‘-minmeta’ argument to create the metadata files and didn’t manually input any of the fields, so I’m not sure what caused the incorrect PhaseEncodingDirection.
Just checked that the PhaseEncodingDirection of the fieldmap metadata is ‘i’, whereas that of the EPI metadata is ‘i-’.
If you could give me any input regarding it, it’d be really helpful. Thank you in advance again!