Summary of what happened:
Hi guys, I’m trying to run ASLPrep through an aslprep.sh script (submitting a job through slurm) but I keep encountering this value error even though I have no desk images - someone suggested it might be an issue with the latest version of ASLPrep, was wondering if anyone has any insight?
Command used (and if a helper script was used, a link to the helper script or the command generated):
and this is my script, I even modified it to bypass the error but it hasn’t worked:
#!/bin/bash
#SBATCH --job-name=ASLprep_sub4028
#SBATCH --output=/data/raulab/aslprep/derivatives/logs/aslprep_%j.out
#SBATCH --error=/data/raulab/aslprep/derivatives/logs/aslprep_%j.err
#SBATCH --nodes=1
#SBATCH --ntasks=1
#SBATCH --cpus-per-task=16
#SBATCH --time=07:00:00
#SBATCH --mail-user=rubana.mursh@gmail.com
#SBATCH --mail-type=FAIL
## cd /data/raulab/aslprep/derivatives/logs
## sbatch /data/raulab/aslprep/aslprep_rubana.sh
# Environment Variables
export FS_LICENSE=/data/raulab/aslprep/config/license.txt
export PYBIDS_INVALID_FILTERS=allow #added to bypass desc error
export APPTAINERENV_PYBIDS_INVALID_FILTERS=allow
export TEMPLATEFLOW_HOME=/data/raulab/aslprep/config/templateflow
export APPTAINERENV_FS_LICENSE=/FreeSurferLicense/license.txt
export APPTAINERENV_TEMPLATEFLOW_HOME=/templateflow
# Node-local work directory
WORKDIR=/scratch/murrub/aslprep_${SLURM_JOB_ID}
mkdir -p "${WORKDIR}"
echo "Using node-local workdir: ${WORKDIR}"
apptainer run --cleanenv \
--env TEMPLATEFLOW_HOME=/templateflow \
--bind /data/raulab/aslprep/preprocessed:/data:ro \
--bind /data/raulab/aslprep/derivatives:/out \
--bind /data/raulab/aslprep/config/license.txt:/freesurfer_license/license.txt \
--bind /data/raulab/aslprep/config/templateflow:/templateflow \
--bind ${WORKDIR}:/work \
/data/raulab/aslprep/aslprep-25.1.0.sif \
/data /out participant \
--participant-label 4028\
--skip_bids_validation \
--output-spaces T1w MNI152NLin2009aSym \
--random-seed 42 \
--n_cpus 16 \
--basil \
--skull-strip-fixed-seed \
--use-syn-sdc \
--fs-license-file /freesurfer_license/license.txt \
--work-dir /work \
--clean-workdir
Version:
25.1.0
Environment (Docker, Singularity / Apptainer, custom installation):
Apptainer, submitted via Slurm on HPC
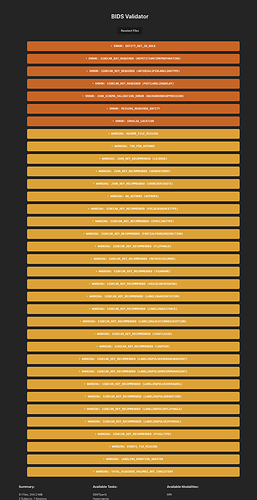
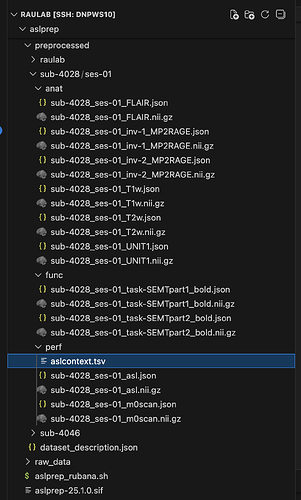
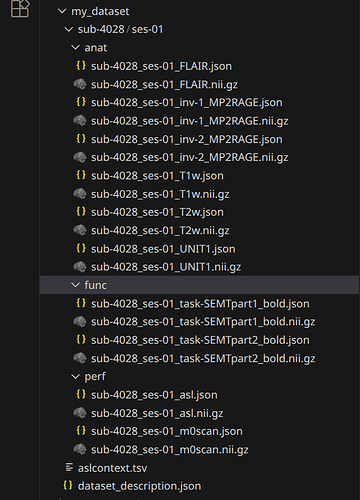
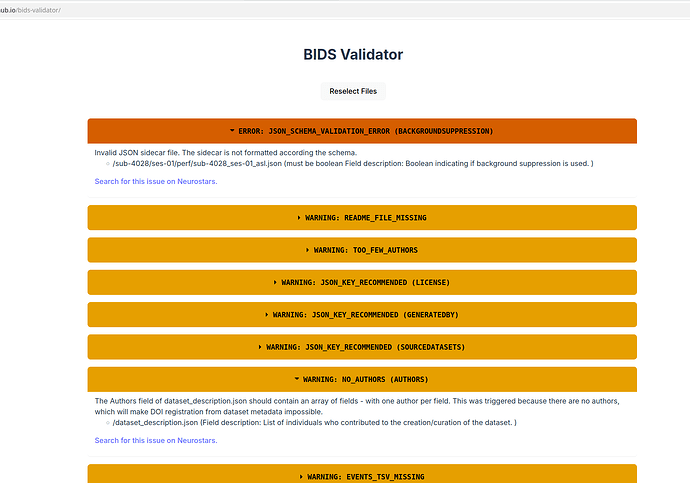
Data formatted according to a validatable standard? Please provide the output of the validator:
PASTE VALIDATOR OUTPUT HERE
Relevant log outputs (up to 20 lines):
this is my error log:
slurmstepd: error: Unable to create TMPDIR [/tmp/user/50084]: Permission denied
slurmstepd: error: Setting TMPDIR to /tmp
INFO: Environment variable SINGULARITY_BINDPATH is set, but APPTAINER_BINDPATH is preferred
WARNING: While bind mounting '/data:/data': destination is already in the mount point list
Process Process-2:
Traceback (most recent call last):
File "/opt/conda/envs/aslprep/lib/python3.11/multiprocessing/process.py", line 314, in _bootstrap
self.run()
File "/opt/conda/envs/aslprep/lib/python3.11/multiprocessing/process.py", line 108, in run
self._target(*self._args, **self._kwargs)
File "/opt/conda/envs/aslprep/lib/python3.11/site-packages/aslprep/cli/workflow.py", line 101, in build_workflow
retval['workflow'] = init_aslprep_wf()
^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/aslprep/lib/python3.11/site-packages/aslprep/workflows/base.py", line 69, in init_aslprep_wf
single_subject_wf = init_single_subject_wf(subject_id)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/aslprep/lib/python3.11/site-packages/aslprep/workflows/base.py", line 575, in init_single_subject_wf
all_estimators, estimator_map = map_fieldmap_estimation(
^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/aslprep/lib/python3.11/site-packages/aslprep/workflows/base.py", line 896, in map_fieldmap_estimation
fmap_estimators = find_estimators(
^^^^^^^^^^^^^^^^
File "/opt/conda/envs/aslprep/lib/python3.11/site-packages/sdcflows/utils/wrangler.py", line 525, in find_estimators
estimator_specs = find_anatomical_estimators(
^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/aslprep/lib/python3.11/site-packages/sdcflows/utils/wrangler.py", line 625, in find_anatomical_estimators
intent = [Path(epi) for epi in layout.get(suffix=suffixes, **query)]
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/aslprep/lib/python3.11/site-packages/bids/layout/layout.py", line 680, in get
raise ValueError(msg + "If you're sure you want to impose "
ValueError: 'desc' is not a recognized entity. If you're sure you want to impose this constraint, set invalid_filters='allow'.