Dear experts,
I am running preprocessing using fmriprep 20.2.3 as usual but this particular subject could not finish recon-all after ~48 hours. For other subjects, it usually takes about ~18-24 hours to finish the recon-all anatomical processing.
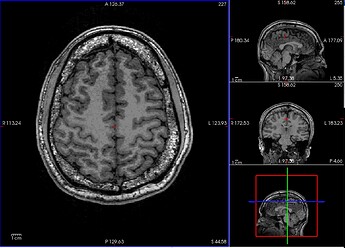
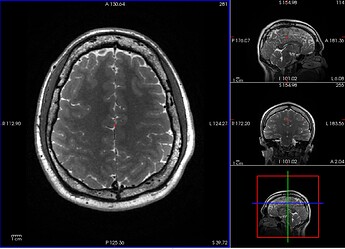
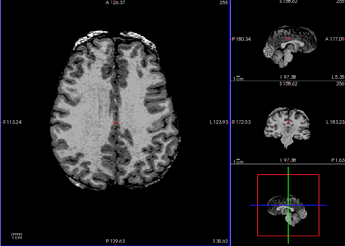
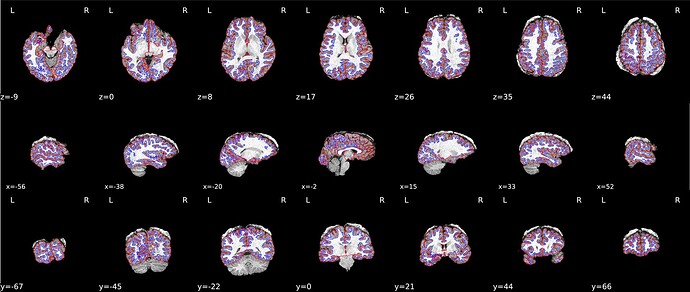
After initial visual inspection, it seems this subject has really thick diploe (T1w and T2w) and part of these bones got detected as brain regions (brainmask.mgz).
T1w
T2w
Brain mask
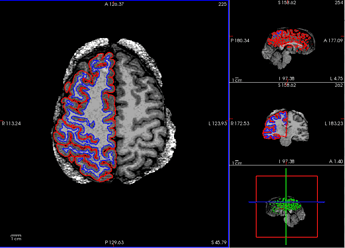
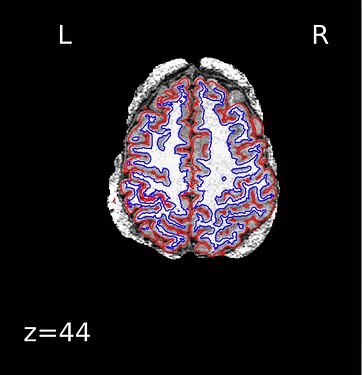
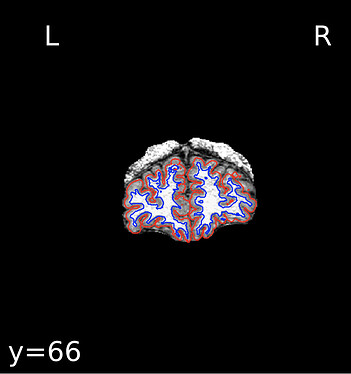
When checking the output from recon-all under the freesurfer subfolder, it seems only the right hemisphere has finished segmentation (below), and the segmentation is mostly right except for tissues near the diploe (the top right side of the brain, shown as the top left). aparc.a2009s+aseg.mgz has not been generated.
For my case, I wonder if you suggest that I give more time to continue running recon-all or any suggestions on how to improve the segmentation? One solution in my mind is to try different Bet options and see which one works the best for removing the diploe.
Thanks for your help in advance!