I have a NIFTI image that I want to register to another coordinate system (I am not sure if registration
is the correct technical term here) using a given affine transformation matrix.
The affine matrix of NIFTI image is
array([[ -1., -0., -0., 0.],
[ -0., -1., -0., 239.],
[ 0., 0., 1., 0.],
[ 0., 0., 0., 1.]])
And the target affine matrix is:
array([[ -1., -0., -0., 0.],
[ -0., 1., -0., 239.],
[ 0., 0., 1., 0.],
[ 0., 0., 0., 1.]])
The difference between the two matrices is the (2,2) element.
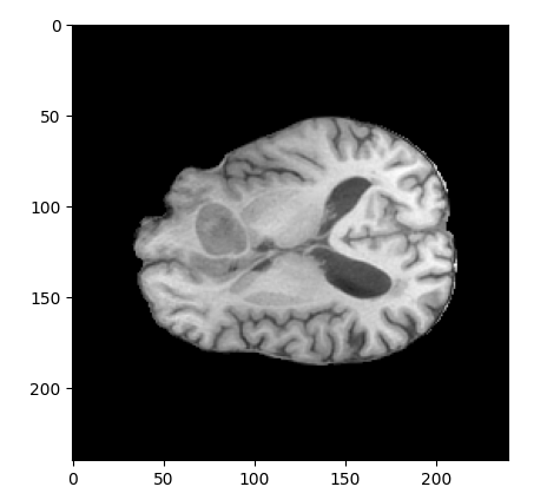
A snippet of a slice in the z-direction for the MRI image is:
And my code to do the registration is:
nifti = nilearn.image.resample_img(nifti, target_affine=target_affine, target_shape=nifti.shape,
interpolation='nearest')
However, the resulting NIFTI image is all zeros. Based on my understanding, the new NIFTI image should be flipped around the second dimension but I think my understanding is off.
- What causes the registered NIFTI image to be zero?
- How to fix this problem?