Hi Nilearn team,
I’m a preclinical fMRI researcher and want to conduct the Nilearn for my data analysis as well as visualization. One major concern is most of the visualization features are too optimized for human research in terms of generating the publishable level of figures while it also provides some flexibility to plot animal data.
So hope to suggest the following features as API to develop some animal-research-friendly features.
- Low-level API guide or instruction to tweak glassy template: Since I’m an animal-oriented researcher, I’m focussing a few species that others not formally do. The code seems only allowed MNI space, and there is no option currently to add additional space for this (hard codded to use only ‘brain_schematics_XX.json’ in ‘glass_brain_files’). So I’m suggesting to tweak the original code as below.
“glass_brain.py”
line 87, add
def _get_json_and_transform(direction, dirname = None):
line 104-105, add
____if dirname is None:
____[indent] dirname = os.path.dirname(os.path.abspath(__file__))
____[indent] dirname = os.path.join(dirname, ‘glass_brain_files’)
line 167, insert
________if ‘dirname’ in kwargs.keys():
____________dirname = kwargs[‘dirname’]
____________del kwargs[‘dirname’]
________else:
____________dirname = None
line 175, add
____json_filename, transform = _get_json_and_transform(direction, dirname)
- The next feature will be decimal position annotation for the smaller brain., Below is my suggestion on updating a code.
“display.py”
line 260, add
____def draw_position(self, size, bg_color, decimal=False, **kwargs):
________if decimal:
____________text = ‘%s=%.{}f’.format(decimal)
________else:
____________text = '%s=%i’
line 262, modify ‘%s=%i’ to text as below
________ax.text(0, 0, text % (self.direction, self.coord),
line 1001, insert
________# use decimal for annotate position
________if ‘decimal’ in kwargs.keys():
____________decimal = kwargs[‘decimal’]
____________del kwargs[‘decimal’]
________else:
____________decimal = False
line 1011, add
________________display_axis.draw_position(size=size, bg_color=bg_color, decimal=decimal, **kwargs)
“img_plotting.py”
line 135, add
____if ‘decimal’ in kwargs.keys():
________decimal = kwargs[‘decimal’]
________del kwargs[‘decimal’]
____else:
________decimal = False
line 194, add
________display.annotate(decimal=decimal)
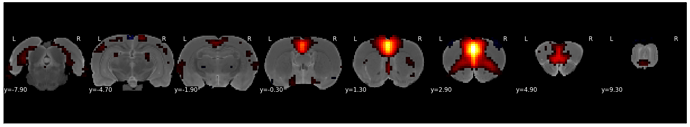
After adding the above code, with passing ‘decimal=2’ on any plotting method, the image will be as below.
I haven’t tested the glass_brain though, so I need the additional test to validate it. but I hope these features could be accepted into the Nilearn for animal researchers and if this looks acceptable, I will generate a pull request according to your development guideline.
Thanks!