I am trying to resample the cerebellum atlas Buckner2011_17Networks_MNI152_FreeSurferConformed1mm_LooseMask.nii.gz to fmriprep output volume ${sub}_task-rest_bold_space-MNI152NLin2009cAsym_brainmask.nii.gz. I was wondering if the following 3 procedures are equivalent (if not what’s the difference?)?
-
mri_vol2vol --mov ${FREESURFER_HOME}/average/Buckner_JNeurophysiol11_MNI152/Buckner2011_17Networks_MNI152_FreeSurferConformed1mm_LooseMask.nii.gz --s ${sub} --targ ${project_path}/fmriprep/${sub}/func/${sub}_task-rest_bold_space-MNI152NLin2009cAsym_brainmask.nii.gz --o ${output_file_path}/Buckner2011_17Networks_MNI152NLin2009cAsym_brainmask.nii.gz --nearest
-
mri_vol2vol --mov ${project_path}/fmriprep/${sub}/func/${sub}_task-rest_bold_space-MNI152NLin2009cAsym_brainmask.nii.gz --s ${sub} --targ ${FREESURFER_HOME}/average/Buckner_JNeurophysiol11_MNI152/Buckner2011_17Networks_MNI152_FreeSurferConformed1mm_LooseMask.nii.gz --o ${output_file_path}/Buckner2011_17Networks_MNI152NLin2009cAsym_brainmask.nii.gz --nearest --inv
-
mri_vol2vol --mov ${FREESURFER_HOME}/average/Buckner_JNeurophysiol11_MNI152/Buckner2011_17Networks_MNI152_FreeSurferConformed1mm_LooseMask.nii.gz --s ${sub} --reg register.dat --o ${output_file_path}/Buckner2011_17Networks_MNI152NLin2009cAsym_brainmask.nii.gz --nearest
where register.dat was generated by the following:
bbregister --s ${sub} --mov ${FREESURFER_HOME}/average/Buckner_JNeurophysiol11_MNI152/Buckner2011_17Networks_MNI152_FreeSurferConformed1mm_LooseMask.nii.gz --int ${project_path}/fmriprep/${sub}/func/${sub}_task-rest_bold_space-MNI152NLin2009cAsym_brainmask.nii.gz --init-coreg --reg register.dat --t2
Thank you for your time!
Regards,
Foucault
I’m not a FreeSurfer expert, but my reading is that:
- 1 and 2 should be equivalent to each other and the correct solution
- 3 includes an unecessary (and potentially incorrectly set up) coregistration step
I personally use nilearn’s resample_to_img for such purposes.
Thanks Chris,
To clarify some concepts, does fmriprep do coregistration with subjects’ T1w while producing ${o_sub}_task-rest_bold_space-MNI152NLin2009cAsym_brainmask.nii.gz?
Wondering why taking the information from a boundary based (co)registration would be a potentially incorrectly setup?
Thanks for the suggestion as well, is resample_to_img similar to the procedure 1 and 2 in my original question? which also implies they do resampling based on identity matrix?
https://mail.nmr.mgh.harvard.edu/pipermail/freesurfer/2018-June/057376.html
(sorry, I asked the same question in Freesurfer mailing list as well)
Yes, but only as an intermediate step. If you want outputs in subject space instead of MNI look for ‘space-T1w’ files.
The fmriprep outputs are already in the MNI space so there is no need to calculated coregistration matrices anew. All you need is to change voxels size and field of view which can be achieved by applying an identity transform.
Yes and yes.
Might be worth replying on the mailing list with a neurostars link for future reference.
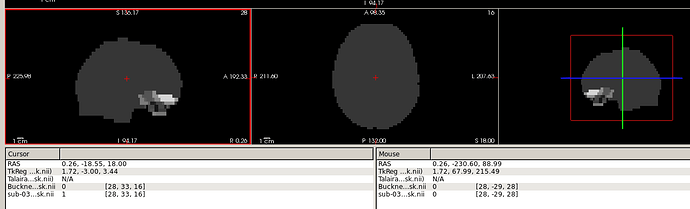
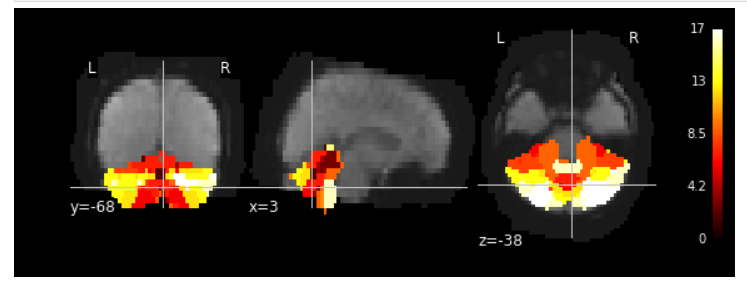
Showing the result of procedure 1 in freeview, these two volume ${sub}_task-rest_bold_space-MNI152NLin2009cAsym_brainmask.nii.gz and ${output_file_path}/Buckner2011_17Networks_MNI152NLin2009cAsym_brainmask.nii.gz did not align well?
So, if I use the voxel indices in Buckner2011_17Networks_MNI152NLin2009cAsym_brainmask.nii.gz to extract roi from sub-03_task-rest_bold_space-MNI152NLin2009cAsym_preproc.nii.gz, I wouldn’t get the correct locations/roi?
Hard to say what I’m suppose to see, but it does look funky! Have you tried resampling with nilearn?
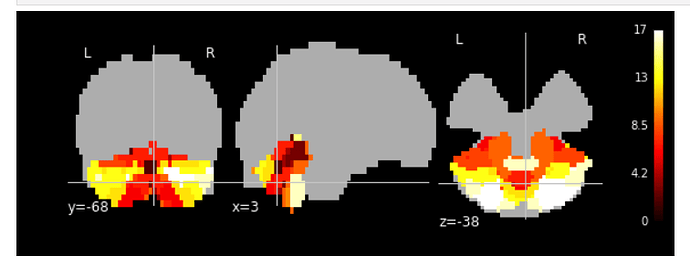
It’s hard to judge from an overlay on top of a brain mask, but this could actually be pretty decent (if this is a cerebellum only atlas). Better use one of the _preproc volumes as a background and show the original atlas on top of an MNI template for reference.
Yes, I think it’s cerebellum only atlas.
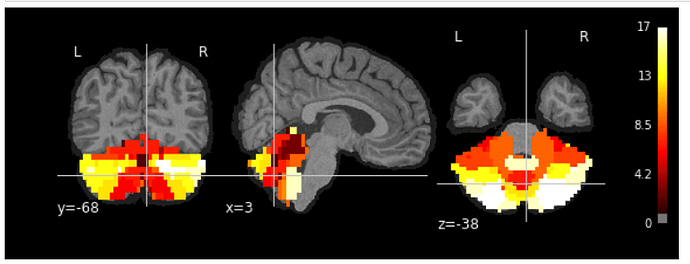
Use _T1w_space-MNI152NLin2009cAsym_preproc.nii.gz as bg_img:
Resampling the cerebellum atlas into _T1w_space-MNI152NLin2009cAsym_preproc.nii.gz, looks even better:
The reason I use _task-rest_bold_space-MNI152NLin2009cAsym_brainmask.nii.gz as a reference for resampling is because that its resolution fits the EPI time series.
My intention is to extract EPI time series (_task-rest_bold_space-MNI152NLin2009cAsym_preproc.nii.gz) based on the cerebellum atlas defined roi. If setting the _T1w_space-MNI152NLin2009cAsym_preproc.nii.gz as resampled reference rather than _task-rest_bold_space-MNI152NLin2009cAsym_brainmask.nii.gz, I would have to resample both the EPI time series and cerebellum atlas into _T1w reference. I am wondering if each additional transformation would distort data a bit?
1 Like
It seems everything worked fine! You are using the right reference. I was only asking you to use “_task-rest_bold_space-MNI152NLin2009cAsym_preproc.nii.gz” as a background for visualisation purposes.
Sorry for the misunderstanding!
In that case, I have to manually extract a single time point form the EPI time series (_task-rest_bold_space-MNI152NLin2009cAsym_preproc.nii.gz) as bg_img right? The nilearn plotting.plot_stat_map module doesn’t support the input as a whole 4d image.
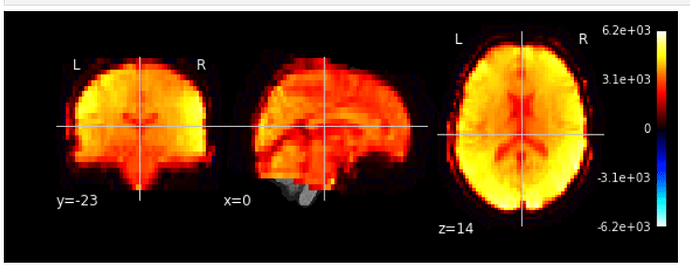
I used mean EPIs as bg_img with cerebellum atlas on top of it. However, the lower part of cerebellum was not covered well.
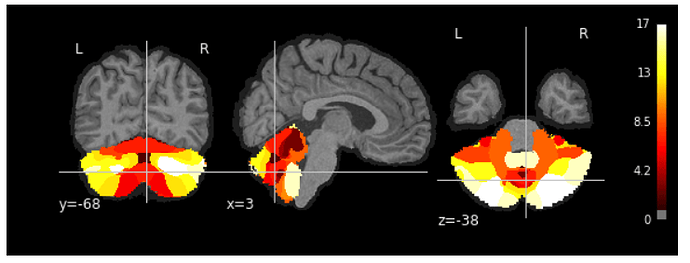
mean EPIs overlayed on top of T1w:
Do you have any suggestions to increase the FOV in the _task-rest_bold_space-MNI152NLin2009cAsym_preproc.nii.gz and _task-rest_bold_space-MNI152NLin2009cAsym_brainmask.nii.gz?
Or the FOV in _preproc.nii.gz and _brainmask.nii.gz is determined by the original EPI FOV?
Thank you for your time!
Is the FOV of the EPI images you see in the FMRIPREP outputs different from the FOV of the input EPI image? It should be the same. Please check the input EPI image of the same participant/task/run.
Most sequences don’t have enough slices to cover all of the brain and the cerebellum.