is it possible to do an ROI SVM analysiswith the use of extracted voi.mat files rather than roi.nii
Hi @Mona-byte,
sorry not a matlab/TDT person, thus not sure how helpful my comments might actually be.
You mean, you want to run some analyzes on already extracted voi.mat files with no access to the underlying/original image data, right? If so, you might have to check how/where TDT extracts values from image files and in what form they are then utilized. IIRC voi.mat contain time series of a given region, eh? Is that an average or does it actually contain the time series of all voxels within a given region? If the former is true then you might not be able to utilize these files, as the original voxel pattern over time has been averaged to reflect one value over time.
HTH, cheers, Peer
Yes, I am running some analysis on already extracted voi.mat files with no access to the underlying/original image data.
How do I check where TDT extracts values from image files
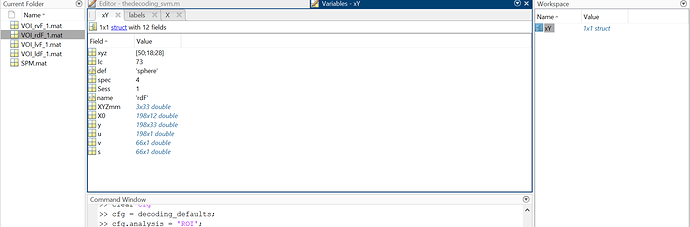
So, my analysis folder has 50 subfolders, each of which contain an spm.mat file and extracted VOI.mat files from 4 roi’s.
Hi Monalisa,
I have no idea what is inside your voi files but I’m just assuming that you know exactly what are the different conditions you want to classify and what are the different runs (or how you want to split up your data into separate chunks). If you don’t have brain volumes it is still possible to run these analyses with TDT. Two ways come to my mind: One is that you use passed_data. Check out our template in the templates folder for that purpose. You would still create an experimental design the normal way if your SPM.mat is intact.
The other way is that you figure out how to write the VOI files to niftis manually, possibly even calling them according to how they were called in the SPM.mat. This would work with default TDT or with the nobetas template. However, looking at the content of your files, this will likely not work easily since the xyz coordinates are only available as XYMmm, and you would manually have to redefine the sphere. So, try it with passed_data.
Best,
Martin
Hi Martin,
Thanks a lot for your response. The VOIs for each subject are the
time-series for each brain region, which have already been prepared. The SPM.mat file has the conditions time series and TR time.
Since I don’t have info on the runs, I think having just 1’s in the chunk should work. Though I’m not sure on how many 1’s to add in the chunk.