Hey Neurostars,
I’ve having a lot of trouble doing a first-level analysis with FEAT of task fMRI data that was pre-processed using fMRIPrep. I have tried scripting out the analysis, after doing a test run in the GUI. However, the errors I have gotten have not been very interpretable. Unfortunately, I don’t see tutorials online that use fMRIPrep output as input for FEAT.
Does anyone have an example design.fsf file I can reference?
Here is an example error I receive. It is with Registration, but I should be able to skip this step since I’m using an output of fMRIPrep.
Registration
/bin/mkdir -p /oak/stanford/groups/nolanw/OCD/Task/first-level-fsl/53879s008/V2++.feat/reg
/home/groups/russpold/software/fsl-6.0.4/bin/mainfeatreg: line 504: /home/groups/russpold/software/fsl-6.0.4/bin/imcp: No such file or directory
/home/groups/russpold/software/fsl-6.0.4/bin/fslmaths /share/software/user/open/fsl/5.0.10/data/standard/MNI152_T1_2mm_brain standard
did not find file: example_func2standard.mat. Generating transform.
/home/groups/russpold/software/fsl-6.0.4/bin/flirt -in example_func -ref standard -out example_func2standard -omat example_func2standard.mat -cost corratio -dof 12 -searchrx -90 90 -searchry -90 90 -searchrz -90 90 -interp trilinear
Image Exception : #63 :: No image files match: example_func
Image Exception : #22 :: Failed to read volume example_func
Error : No image files match: example_func
Failed to read volume example_func
Error : No image files match: example_func
/home/groups/russpold/software/fsl-6.0.4/bin/slicer example_func2standard standard -s 2 -x 0.35 sla.png -x 0.45 slb.png -x 0.55 slc.png -x 0.65 sld.png -y 0.35 sle.png -y 0.45 slf.png -y 0.55 slg.png -y 0.65 slh.png -z 0.35 sli.png -z 0.45 slj.png -z 0.55 slk.png -z 0.65 sll.png ; /home/groups/russpold/software/fsl-6.0.4/bin/pngappend sla.png + slb.png + slc.png + sld.png + sle.png + slf.png + slg.png + slh.png + sli.png + slj.png + slk.png + sll.png example_func2standard1.png ; /home/groups/russpold/software/fsl-6.0.4/bin/slicer standard example_func2standard -s 2 -x 0.35 sla.png -x 0.45 slb.png -x 0.55 slc.png -x 0.65 sld.png -y 0.35 sle.png -y 0.45 slf.png -y 0.55 slg.png -y 0.65 slh.png -z 0.35 sli.png -z 0.45 slj.png -z 0.55 slk.png -z 0.65 sll.png ; /home/groups/russpold/software/fsl-6.0.4/bin/pngappend sla.png + slb.png + slc.png + sld.png + sle.png + slf.png + slg.png + slh.png + sli.png + slj.png + slk.png + sll.png example_func2standard2.png ; /home/groups/russpold/software/fsl-6.0.4/bin/pngappend example_func2standard1.png - example_func2standard2.png example_func2standard.png; /bin/rm -f sl?.png example_func2standard2.png
Image Exception : #63 :: No image files match: example_func2standard
Image Exception : #22 :: Failed to read volume example_func2standard
Error : No image files match: example_func2standard
Cannot open sla.png for reading
sh: line 1: 9833 Segmentation fault /home/groups/russpold/software/fsl-6.0.4/bin/pngappend sla.png + slb.png + slc.png + sld.png + sle.png + slf.png + slg.png + slh.png + sli.png + slj.png + slk.png + sll.png example_func2standard1.png
Cannot open example_func2standard1.png for reading
sh: line 1: 9837 Segmentation fault /home/groups/russpold/software/fsl-6.0.4/bin/pngappend example_func2standard1.png - example_func2standard2.png example_func2standard.png
Error in slicer input, exiting...
Howdy @Ruben_Krueger ,
First, you will want to make sure your data is in bids format.
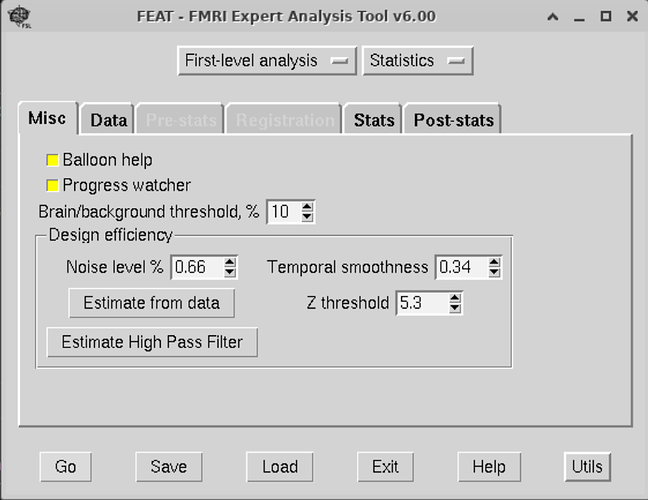
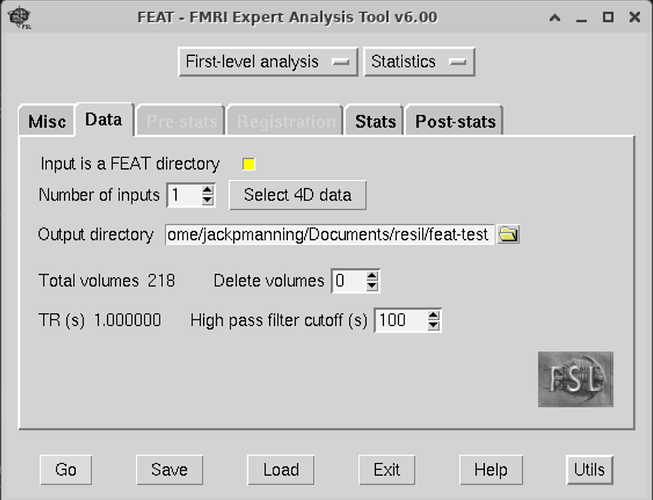
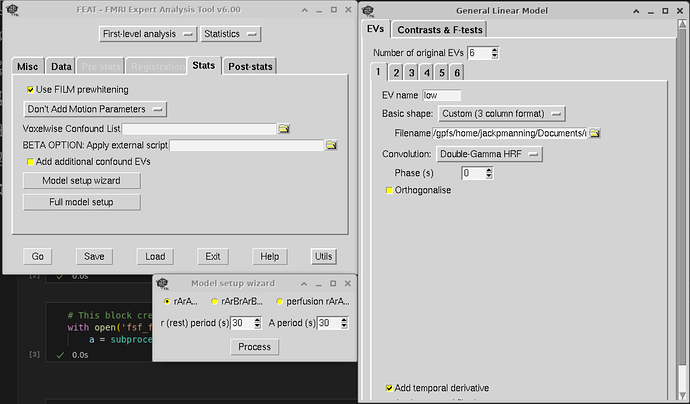
Second, you will want to create a template.fsf file. To do this, you will have to go through the FSL gui with one scan, and save the fsf file. For this step, both mumfordbrainstats and andy’s brain blog have videos on this. The template.fsf file will have to be tailored to your data set, so you won’t be able to just cop/paste someone else’s. Since you have already preprocessed, you will want to select ‘First-level analysis’ and ‘Statistics’ at the top of FEAT.
Third, copy/paste and rename your .fsf file and put searchable strings into it that you can search/replace with with a python script (or whatever your preferred language is). These strings will go in places that would change each for each iteration of a loop that you would run to go through all of the subjects. For example, I put SUBNUM in my template.fsf file, and created a dictionary in python where SUBNUM takes on the value of the subject identifier during each loop iteration.
In my experience, this process is a lot more complicated than it originally seems. I am currently going through the same thing with my data. I just finished my first level analysis in fsl using fmirprepped data.
Please check out this git repo that I created for the project I am working on. I have not yet added any description on how to use the fsl code. Please not that these are specific to my experimental setup (sternberg protocol). The files in fsl-code are:
template.fsf: the template I used for search/replacing
make_fsf_lev_1.py: python script to create the fsf files for each run
run_fsl_python.ipynb: jupyter notebook using python to run fsl for each .fsf file I created.
I just finished doing this step last week, so hopefully this is helpful. These are screenshots for setting up the feat gui:
edit: I added the github link
Hey @jackpmanning,
Thank you so much for the detailed response. I will try adapting my design.fsf based on the template.fsf in the Github repo.
Hello,
I have completed the pre-processing of my n-back task-based fMRI data using fmriprep as you. Now, I want to perform the feat analysis with FSL, incorporating my task characteristics. However, I cannot find any resources regarding the expected outputs. Does anyone know clearly what outputs I should expect after running the FEAT analysis with FSL?
Hi Beatriz,
I assume you are attempting to perform the first-level analysis. The output files will depend on your settings in FEAT. The FEAT wiki has a good explanation of the expected output files:
https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FEAT/UserGuide#FEAT_Output
Generally, the generated “.feat” directories from the first level analysis will be used as input to the higher level (i.e., group level analysis).