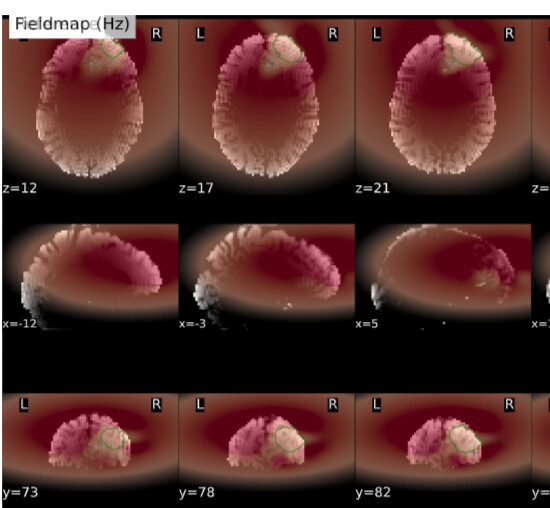
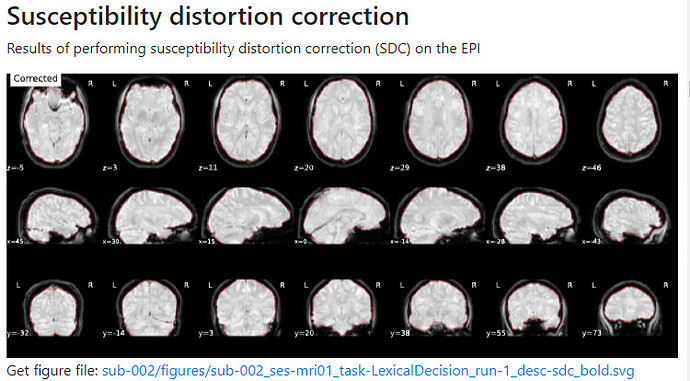
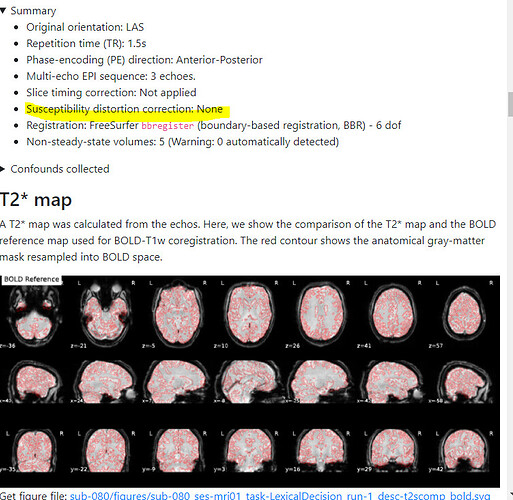
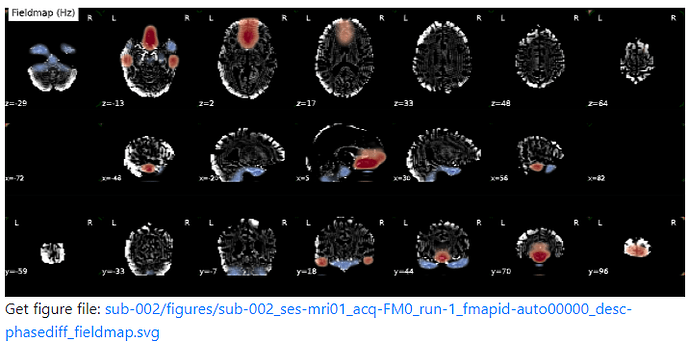
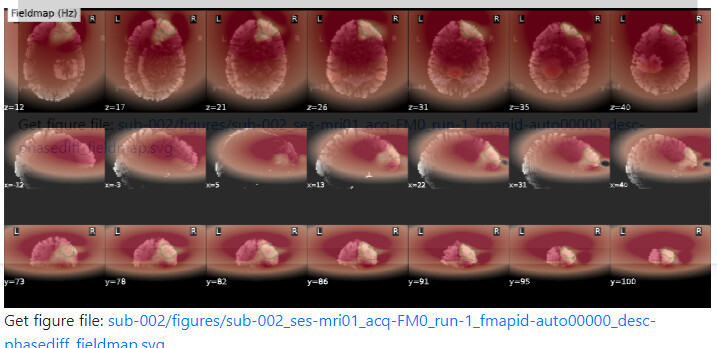
Summary of what happened:
For a couple of data sets SDC does not seem to work using fmriprep v22.1.0/22.1.1. For most of the participants with the same data structure, the pipeline seems to work (~180 datasets) but for a few I get the same error message regarding bs_filter node giving errors. My fieldmap has 2 magnitude files and 1 phase difference image. While executing fmriprep v22.1.0, we noticed a weird distortion of the image, and thus we BET extracted the mag and the phase diff images using funtions in FSL before applying fmriprep. The datasets in question did not give error messages when v20.2.3 was used.
Command used (and if a helper script was used, a link to the helper script or the command generated):
fmriprep_sub.py /project/******/MRI/bids_v3.7.4 -o /project/3011099.01/MRI/fmriprep/fmriprep_v22.1.1_LD -m 84000 -p 064 -a "--skip_bids_validation -t LexicalDecision --use-aroma --aroma-melodic-dimensionality 100 --ignore slicetiming --output-spaces MNI152NLin6Asym:res-2 T1w --dummy-scans 5"
fmriprep_sub.py is a wrapper python script to run fmriprep
Version:
fmriprep v22.1.0 and v22.1.1
Environment (Docker, Singularity, custom installation):
singularity container
Data formatted according to a validatable standard? Please provide the output of the validator:
bids-validator did not give any error messages
Relevant log outputs (up to 20 lines):
Error message:
Node Name: fmriprep_22_1_wf.single_subject_064_wf.fmap_preproc_wf.wf_FM0.bs_filter
File: `/project/********/MRI/fmriprep_v22.1.0_LD/sub-064/log/20221220-123516_9bd453bc-f81c-4066-8e0d-2b9042cb7fdf/crash-20221220-125616-atstak-bs_filter-3692ad96-2797-4aa7-b09a-a0564718a548.txt`
Working Directory: `/scratch/atstak/48315571.dccn-l029.dccn.nl/sub-064/fmriprep_22_1_wf/single_subject_064_wf/fmap_preproc_wf/wf_FM0/bs_filter`
Inputs:
* bs_spacing: `[(100.0, 100.0, 40.0), (16.0, 16.0, 10.0)]`
* debug: `False`
* extrapolate: `True`
* in_data:``
* in_mask:``
* recenter: `mode`
* ridge_alpha: `0.01`
* zooms_min: `4.0`
Screenshots / relevant information:
From the o-file of the failed dataset:
230125-02:12:51,516 nipype.workflow WARNING:
[Node] Error on "fmriprep_22_1_wf.single_subject_064_wf.fmap_preproc_wf.wf_FM0.bs_filter" (/scratch/atstak/48453721.dccn-l029.dccn.nl/sub-064/fmriprep_22_1_wf/single_subject_064_wf/fmap_preproc_wf/wf_FM0/bs_filter)
230125-02:12:52,155 nipype.interface INFO:
Approximating B-Splines grids (5x5x6, and 15x15x15 [knots]) on a grid of 52x52x32 (86528) voxels, of which 83824 fall within the mask.
230125-02:12:53,135 nipype.workflow ERROR:
Node bs_filter failed to run on host dccn-c057.dccn.nl.
230125-02:12:53,143 nipype.workflow ERROR:
....
.....
.....
....
230125-07:56:47,495 nipype.workflow CRITICAL:
fMRIPrep failed: Traceback (most recent call last):
File "/opt/conda/lib/python3.9/site-packages/nipype/pipeline/plugins/multiproc.py", line 67, in run_node
result["result"] = node.run(updatehash=updatehash)
File "/opt/conda/lib/python3.9/site-packages/nipype/pipeline/engine/nodes.py", line 527, in run
result = self._run_interface(execute=True)
File "/opt/conda/lib/python3.9/site-packages/nipype/pipeline/engine/nodes.py", line 645, in _run_interface
return self._run_command(execute)
File "/opt/conda/lib/python3.9/site-packages/nipype/pipeline/engine/nodes.py", line 771, in _run_command
raise NodeExecutionError(msg)
nipype.pipeline.engine.nodes.NodeExecutionError: Exception raised while executing Node bs_filter.
Traceback:
Traceback (most recent call last):
File "/opt/conda/lib/python3.9/site-packages/nipype/interfaces/base/core.py", line 398, in run
runtime = self._run_interface(runtime)
File "/opt/conda/lib/python3.9/site-packages/sdcflows/interfaces/bspline.py", line 186, in _run_interface
data -= np.squeeze(mode(data[mask]).mode)
ValueError: operands could not be broadcast together with shapes (52,52,32) (0,) (52,52,32)
230125-07:56:50,519 cli ERROR:
Preprocessing did not finish successfully. Errors occurred while processing data from participants: 064 (1). Check the HTML reports for details.
*The crash report states:*
Node: fmriprep_22_1_wf.single_subject_064_wf.fmap_preproc_wf.wf_FM0.bs_filter
Working directory: /scratch/atstak/48315089.dccn-l029.dccn.nl/sub-064/fmriprep_22_1_wf/single_subject_064_wf/fmap_preproc_wf/wf_FM0/bs_filter
Node inputs:
bs_spacing = [(100.0, 100.0, 40.0), (16.0, 16.0, 10.0)]
debug = False
extrapolate = True
in_data = <undefined>
in_mask = <undefined>
recenter = mode
ridge_alpha = 0.01
zooms_min = 4.0
Traceback (most recent call last):
File "/opt/conda/lib/python3.9/site-packages/nipype/pipeline/plugins/multiproc.py", line 67, in run_node
result["result"] = node.run(updatehash=updatehash)
File "/opt/conda/lib/python3.9/site-packages/nipype/pipeline/engine/nodes.py", line 527, in run
result = self._run_interface(execute=True)
File "/opt/conda/lib/python3.9/site-packages/nipype/pipeline/engine/nodes.py", line 645, in _run_interface
return self._run_command(execute)
File "/opt/conda/lib/python3.9/site-packages/nipype/pipeline/engine/nodes.py", line 771, in _run_command
raise NodeExecutionError(msg)
nipype.pipeline.engine.nodes.NodeExecutionError: Exception raised while executing Node bs_filter.
Traceback:
Traceback (most recent call last):
File "/opt/conda/lib/python3.9/site-packages/nipype/interfaces/base/core.py", line 398, in run
runtime = self._run_interface(runtime)
File "/opt/conda/lib/python3.9/site-packages/sdcflows/interfaces/bspline.py", line 186, in _run_interface
data -= np.squeeze(mode(data[mask]).mode)
ValueError: operands could not be broadcast together with shapes (52,52,32) (0,) (52,52,32)