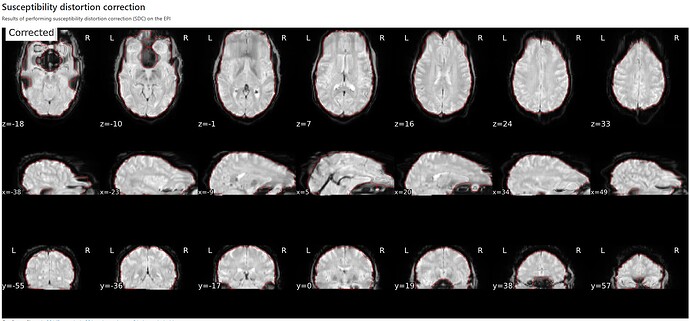
Summary of what happened:
Hi everyone, I am trying to run fMRIprep on some pilot data to establish a pipeline. When performing SDC via the phase-difference technique, I am getting an output that is severely distorted in the frontal region. Pictures below:
Command used (and if a helper script was used, a link to the helper script or the command generated):
apptainer run --cleanenv \
-B "${FS_LICENSE_FILE}":"${FS_LICENSE_FILE}" \
-B /project/6003371:/project/6003371 \
-B /localscratch:/localscratch \
-B /cvmfs:/cvmfs \
-B /project:/project \
-B /scratch:/scratch \
-B /local:/local \
"$SIF" \
"$BIDS_DIR" "$OUT_DIR" participant \
--participant_label sub-001 \
--nthreads 8 --n_cpus 8 --mem_mb 32000 --omp-nthreads 8 \
-w "$WORK_DIR" \
--fs-license-file "$FS_LICENSE_FILE" \
--skull-strip-t1w force \
--output-spaces MNI152NLin2009cAsym fsaverage fsnative \
--fd-spike-threshold 1.5 \
--skip_bids_validation
Version:
23.1.4
Environment (Docker, Singularity / Apptainer, custom installation):
Using Apptainer through Compute Canada
Data formatted according to a validatable standard? Please provide the output of the validator:
#Data was converted using dcm2bids
Relevant log outputs (up to 20 lines):
#fMRIprep finished successfully
Screenshots / relevant information:
Phase-difference estimation SDC:
We also collected fmaps to try the PEPOLAR technique as well, the distortion is less but still bad. However, registration looks good when SDC is not run. The unfortunate thing is that we’ve run fMRIprep on another dataset in the lab, and the current problem still occurs. I suspect it may be something with the dcms2bids conversion?
Here is the information in the phasediff.json:
{
"Modality": "MR",
"MagneticFieldStrength": 3,
"ImagingFrequency": 123.219469,
"Manufacturer": "Siemens",
"ManufacturersModelName": "MAGNETOM Prisma Fit",
"InstitutionName": "###",
"InstitutionAddress": "###",
"DeviceSerialNumber": "###",
"StationName": "###",
"BodyPartExamined": "BRAIN",
"PatientPosition": "HFS",
"ProcedureStepDescription": "###",
"SoftwareVersions": "syngo MR XA60",
"MRAcquisitionType": "2D",
"SeriesDescription": "gre_field_mapping",
"ProtocolName": "gre_field_mapping",
"ScanningSequence": "GR",
"SequenceVariant": "SP",
"PulseSequenceName": "*fm2d2r",
"ImageType": [
"ORIGINAL",
"PRIMARY",
"P",
"NONE",
"PHASE"
],
"ImageTypeText": [
"ORIGINAL",
"PRIMARY",
"P",
"DIS2D"
],
"NonlinearGradientCorrection": true,
"SeriesNumber": 30,
"AcquisitionTime": "12:43:27.562500",
"AcquisitionNumber": 1,
"SliceThickness": 3,
"SpacingBetweenSlices": 3.75,
"EchoNumber": 2,
"EchoTime": 0.00738,
"RepetitionTime": 0.5,
"MTState": false,
"FlipAngle": 60,
"EchoTime1": 0.00492,
"EchoTime2": 0.00738,
"PartialFourier": 1,
"BaseResolution": 80,
"ShimSetting": [
5189,
-6303,
-3968,
354,
117,
220,
-149,
-103
],
"TxRefAmp": 295.756,
"PhaseResolution": 1,
"ReceiveCoilName": "OpenFace_28",
"ReceiveCoilActiveElements": "Rx1",
"CoilString": "OpenFace_28",
"PulseSequenceDetails": "%SiemensSeq%\\gre_field_mapping",
"CoilCombinationMethod": "Sum of Squares",
"MatrixCoilMode": "SENSE",
"PercentPhaseFOV": 100,
"PercentSampling": 100,
"PhaseEncodingSteps": 80,
"FrequencyEncodingSteps": 80,
"AcquisitionMatrixPE": 80,
"ReconMatrixPE": 80,
"AcquisitionDuration": 79.487,
"PixelBandwidth": 291,
"DwellTime": 2.15e-05,
"PhaseEncodingDirection": "j-",
"SliceTiming": [
0.2475,
0,
0.26,
0.01,
0.27,
0.02,
0.28,
0.0325,
0.2925,
0.0425,
0.3025,
0.0525,
0.3125,
0.065,
0.325,
0.075,
0.335,
0.085,
0.3475,
0.0975,
0.3575,
0.1075,
0.3675,
0.1175,
0.38,
0.13,
0.39,
0.14,
0.4,
0.15,
0.4125,
0.1625,
0.4225,
0.1725,
0.4325,
0.1825,
0.445,
0.195,
0.455,
0.205,
0.465,
0.215,
0.4775,
0.2275,
0.4875,
0.2375
],
"ImageOrientationPatientDICOM": [
0.998441,
-0.0558215,
0,
0.0558215,
0.998441,
0
],
"InPlanePhaseEncodingDirectionDICOM": "COL",
"BidsGuess": [
"fmap",
"_acq-fm2_phasediff"
],
"ConversionSoftware": "dcm2niix",
"ConversionSoftwareVersion": "v1.0.20240202",
"Dcm2bidsVersion": "3.2.0",
"IntendedFor": [
"func/sub-001_task-movie_run-01_bold.nii.gz",
"func/sub-001_task-physics_run-02_bold.nii.gz",
"func/sub-001_task-tool_run-03_bold.nii.gz",
"func/sub-001_task-movie_run-02_bold.nii.gz",
"func/sub-001_task-tool_run-01_bold.nii.gz",
"func/sub-001_task-tool_run-04_bold.nii.gz",
"func/sub-001_task-physics_run-01_bold.nii.gz",
"func/sub-001_task-tool_run-02_bold.nii.gz"
]
}
Grateful for any help! Thank you!