Hi there,
I’m trying to run a second-level group comparison using Nilearn for a task-based fMRI paradigm. At the first level, I specified two contrasts for each subject: Stim1 > Stim2 and Stim2 > Stim1. Now, I want to compare these contrasts across three groups.
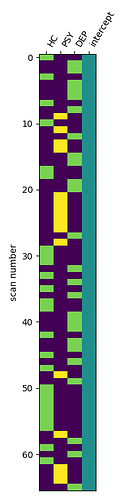
For the second-level GLM, I included dummy-coded group variables along with an intercept. To compare group A vs. group B, I set the contrast to [1, -1, 0, 0]. I also examined the main effect by dropping the group variables and using only the intercept. Additionally, for exploratory purposes, I looked at the effect of individual groups, e.g., using the contrast [1, 0, 0, 0] for group A.
However, I’m getting identical results for all group comparisons, which doesn’t seem correct. I suspect that the intercept might be coded differently than I expect, possibly affecting how the contrasts are interpreted. I would appreciate any advice on what might be going wrong or how to properly set up the contrasts.
Many thanks in advance!
Annkathrin