Hello,
I want to do statistical test for my searchlight results, and I found it seems that the SecondLevelModel/non_parametric_inference/permuted_ols functions are similar.
Firstly I did a one-sample test by a second-level fMRI model.
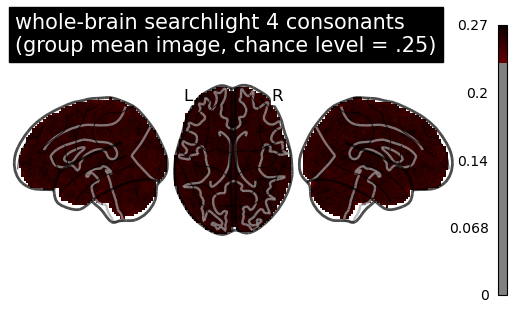
Here is the mean image of the searchlight.scores_ across participants (chance level = .25):
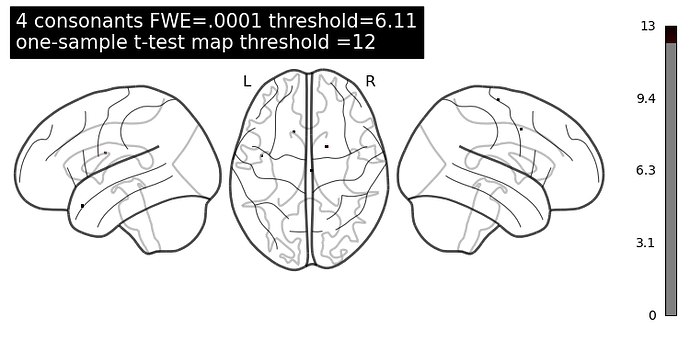
and here is the result using SecondLevelModel for a one-sample test:
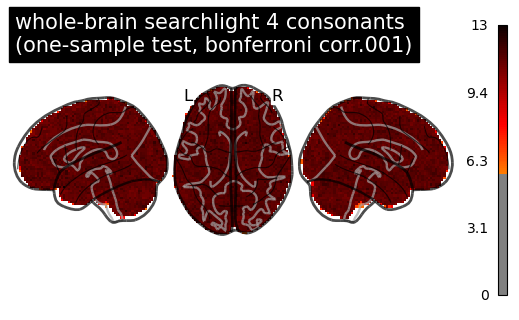
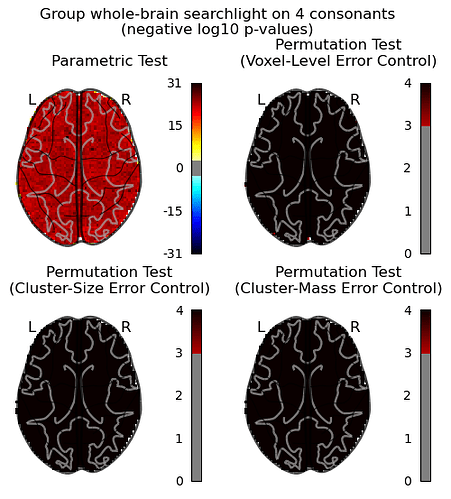
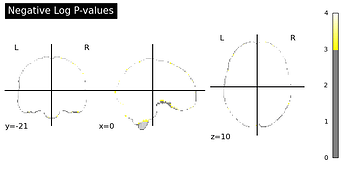
then I computed a corrected p-values with a permutation test following the example by non_parametric_inference(), and the argument design_matrix is the same one as I used in SecondLevelModel, so intrcept = 0.25.
I think something is obviously off here. For the glass brain “Voxel-Level Error Control” I set the threshold=None as mentioned in the example for running the voxel-level correction.
Instead, if I just threshold the result of one-sample test, I can at least see the voxels with the highest classification score.
Question:
- Am I right to do the one-sample test with the permutation test here?
- The input images here is different from the example that they are an array of classification accuracy score, so did I miss something here?
- I also take a look of another example of massively univariate analysis, but I don’t understand how to do this with the searchlight results in which we don’t have labels anymore.