I would like to do things with the cifti beta maps in HCP using python.
But first I want to make sure that I know where I am in the brain, that is, how do the grayordinates map onto different structures.
Tl;dr: How do I subset the grayordinates so that I have the left and right hemisphere vertices in python?
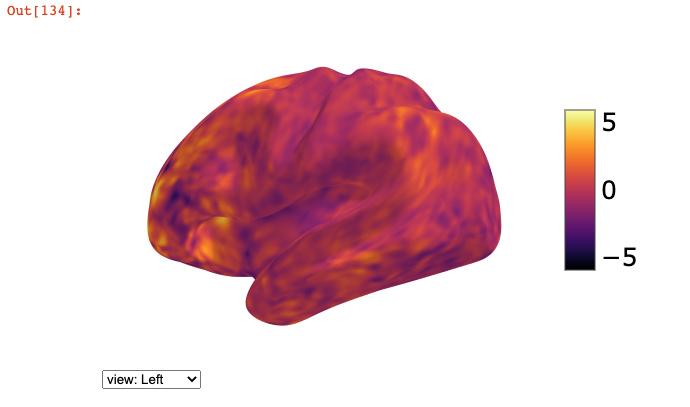
Let’s say I just want to view the left hemisphere beta map overlaid onto the group average mesh. I have confirmed that they are both 32k_fs_LR mesh.
beta map
tfMRI_GAMBLING_hp200_s2_level2_MSMAll.feat/GrayordinatesStats/cope1.feat/pe1.dtseries.nii
mesh:
S1200.L.inflated_MSMAll.32k_fs_LR.surf.gii
Using an example derived from the nibabel cifit examples, I am doing the following:
import numpy as np
import nibabel as nib
from nilearn import plotting
# load image
img = nib.cifti2.load('pe1.dtseries.nii')
# get brain model for left cortex
bml = [f for f in img.header.matrix.get_index_map(1).brain_models if f.brain_structure == 'CIFTI_STRUCTURE_CORTEX_LEFT']
# get image offset (not really relevant for left cortex, but will be for right)
index_offset = bml[0].index_offset
# get left hemi vertex indices
vertexIndicesInDataLeft = [i+index_offset for i in bml[0].vertex_indices]
# count the number of vertices
len(vertexIndicesInDataLeft)
29696 # why is this not 32492?
# get the beta values
beta_map = img.get_fdata()
# subset left vertices
left_map = beta_map[0,vertexIndicesInDataLeft]
# create zero pad to expand to 32k
map_full = np.zeros((1,32492))
# put beta values onto full map
map_full[0,vertexIndicesInDataLeft] = left_map
# load mesh
mesh = nib.load('S1200.L.inflated_MSMAll.32k_fs_LR.surf.gii')
# plot
plotting.view_surf(mesh.agg_data(), map_full, symmetric_cmap=False, cmap='inferno')
The above code gives me a trippy brain. I suspect it has something to do with how I’m indexing the left hemi’s vertices.

If I use the connectome workbench to separate the cifti into a left hemi gifti, it works fine.
wb_command -cifti-separate pe1.dtseries.nii COLUMN -metric CORTEX_LEFT pe1.L.32k_fs_LR.func.gii
then
mesh = nib.load('S1200.L.inflated_MSMAll.32k_fs_LR.surf.gii')
beta_map = nib.load('pe1.L.32k_fs_LR.func.gii').agg_data()
plotting.view_surf(mesh.agg_data(), beta_map, symmetric_cmap=False, cmap='inferno')