I’ve implemented a nipype pipeline to take our data from BIDS → fmriprep → preprocessing → analysis, but I’ve encountered a strange result that I’m trying to understand.
We are using a basic navigation task where we expect the navigation network should be activated.
I’ve run fmriprep and I’m using nipype and FSL (implemented in nipype, version 1.4.2) to clean and analyze the results.
We do get the navigation network activated (hooray!), but only using the uncleaned data. (That is, the data that fmriprep outputs into derivatives: e.g., sub-dspfmri11002_task-dspfmri_run-1_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz
Of course, we’d also like to clean our data. I’m doing so using this script:
# Kernel = UFRC Python-3.8
# TODO:
# Meta-data for the preprocessed data says slicetiming is true, but it is definitely not. What's up? Future Steve.
# To run this:
# See clean_fmriprep_data.sh
from nilearn import image as nimg
import nibabel as nib
import bids # https://bids-standard.github.io/pybids/generated/bids.layout.BIDSLayout.html
import os
import matplotlib.pyplot as plt
import numpy as np
import argparse
import pandas as pd
from nipype.interfaces import fsl
# Arguments to take from the bash script
parser = argparse.ArgumentParser(description="Give me a path to your fmriprep output")
parser.add_argument(
"-i",
"--fmriprepdir",
default=None,
type=str,
help="This is the full path to your fmriprep dir",
)
parser.add_argument(
"-o",
"--outputdir",
default=None,
type=str,
help="Where output data should be saved",
)
parser.add_argument(
"-s", "--subj", default=None, type=str, help="subject ID (without leading 'sub')"
)
parser.add_argument("-t", "--task", default=None, type=str, help="Name of the task")
parser.add_argument(
"--space",
default="MNI152NLin2009cAsym",
type=str,
help="Space for the data to be loaded in",
)
parser.add_argument(
"--highpass", default=0.009, type=float, help="Value for highpass filter"
)
parser.add_argument(
"--lowpass", default=0.08, type=float, help="Value for lowpass filter"
)
parser.add_argument(
"--fwhm", default=5, type=float, help="Value for smoothing kernel size"
)
parser.add_argument(
"-d", "--debug", help="Print debug logs", required=False, action="store_true"
)
args = parser.parse_args()
fmriprep_path = args.fmriprepdir
sub = args.subj
output_path = args.outputdir + os.sep + "sub-" + sub
task = args.task
space = args.space
HIGH_PASS = args.highpass
LOW_PASS = args.lowpass
FWHM = args.fwhm
verbose = args.debug
def get_output_name(input_name, output_path, output_suffix):
# Trim off the nifti suffix
input_name = input_name.split(os.sep)[-1].split(".")[0]
# Absolute path and new name
output_name = output_path + os.sep + input_name + "_" + output_suffix
return output_name
# Check for output folder
if not os.path.isdir(output_path):
os.makedirs(output_path)
print("created folder : ", output_path)
else:
print(output_path, "folder already exists.")
# This tells us where the BIDS directory is.
layout = bids.BIDSLayout(fmriprep_path, validate=False, config=["bids", "derivatives"])
# Define our func, mask, confounds files
func_files = layout.get(
subject=sub,
task=task,
desc="preproc",
suffix="bold",
space=space,
extension="nii.gz",
return_type="file",
)
mask_files = layout.get(
subject=sub,
task=task,
desc="brain",
suffix="mask",
space=space,
extension="nii.gz",
return_type="file",
)
confound_files = layout.get(
subject=sub, task=task, desc="confounds", extension="tsv", return_type="file"
)
for i in range(len(func_files)):
print("----------------------")
print("Beginning run " + str(i + 1))
func_file = func_files[i]
mask_file = mask_files[i]
confound_file = confound_files[i]
if verbose:
print(f"Func file: {func_file}")
print(f"Mask file: {mask_file}")
print(f"Confound file: {confound_file}")
# Make sure everything is the same run
run = "run-" + str(i + 1)
assert run in func_file and run in mask_file and run in confound_file
# Grab TR automatically from the meta data
meta_data_func = layout.get_metadata(func_file)
T_R = meta_data_func["RepetitionTime"]
if verbose:
print(f"TR found as {T_R}")
# From David Smith:
# Also, here: https://www.sciencedirect.com/science/article/pii/S1053811917310972?via%3Dihub
# Delimiter is \t --> tsv is a tab-separated spreadsheet
confound_df = pd.read_csv(confound_file, delimiter="\t")
# Motion outliers (fd threshold from fmriprep defaults)
# NOTE: This is to be used for CENSORING - NOT for outlier removal.
motion_outliers = [col for col in confound_df.columns if 'motion_outlier' in col]
# In case of dummy scans
#non_steady_state = [col for col in confound_df.columns if col.startswith('non_steady_state')]
# PCA first 5 components
# a_comp_cor = ['a_comp_cor_00','a_comp_cor_01','a_comp_cor_02','a_comp_cor_03','a_comp_cor_04','a_comp_cor_05'] # use aCompCor instead
#cosine = [col for col in confound_df.columns if col.startswith('cosine')]
# Motion regressors
motion_trans = [col for col in confound_df.columns if col.startswith('trans_')]
motion_rot = [col for col in confound_df.columns if col.startswith('rot_')]
#motion = ['trans_x','trans_y','trans_z','rot_x','rot_y','rot_z']
filter_col=np.concatenate([motion_outliers,motion_trans,motion_rot])
#filter_col=np.concatenate([motion])
confound_df = confound_df[filter_col]
# Fill nans with 0's
confound_df = confound_df.fillna(0)
if verbose:
print(f"Confounds: {confound_df.columns}")
confounds_matrix = confound_df.values
confounds_output_file = get_output_name(confound_file, output_path, "_trimmed.csv")
np.savetxt(confounds_output_file,confounds_matrix,delimiter=",")
# Load in functional data
raw_func_img = nimg.load_img(func_file)
# Make sure these are the same length.
assert raw_func_img.shape[3] == confounds_matrix.shape[0]
# Clean!
clean_img = nimg.clean_img(
raw_func_img,
confounds=confounds_matrix,
detrend=True,
standardize=True,
low_pass=LOW_PASS,
high_pass=HIGH_PASS,
t_r=T_R,
mask_img=mask_file,
)
# Smooth!
clean_img_smooth = nimg.smooth_img(clean_img, FWHM)
# Write out!
output_file = get_output_name(func_file, output_path, "clean_smooth_mask.nii.gz")
clean_img_smooth.to_filename(output_file)
fsl.maths.ApplyMask(
in_file=output_file, mask_file=mask_file, out_file=output_file
).run()
print(f"Clean and smooth file saved {output_file}")
With everything else the same before and after the above script runs, the contrast between two of our conditions (navigation and locomotion) results in anti-correlated t-statistics. (the correlation coefficient between all voxels in the clean vs. raw data is r = -.31.) The predicted direction (https://neurosynth.org/analyses/terms/navigation/) is consistent with the negative value of the contrast for the cleaned data, but the positive value of the contrast for the raw data. We’ve also run that script with several different combinations of predictors (including just the main 6 motion parameters).
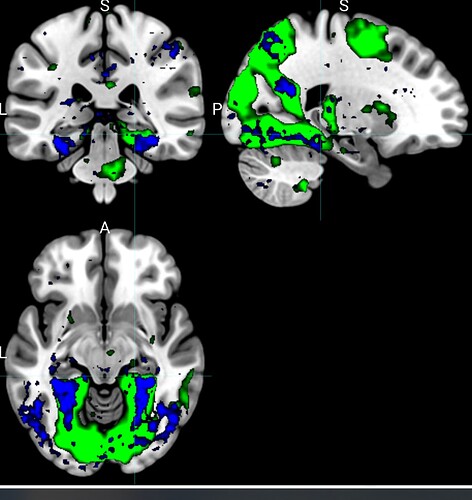
You can see the overlapping brain maps here, noting that the green (clean data) runs between t = -3.0 to t = -.5 ;the blue (raw fmriprep output) runs between 2.0 and 4.0
Curiously, the main effect of condition compared to instructions / fixation is in the same direction for both of these contrasts. I can attach the design.fsf file if that’s helpful as well.
I’d appreciate any troubleshooting tips and can provide more info as helpful.