Hello, I want to run single-subject ICA on preprocessed images from the Human Connectome Project.
Is there any way I can do this using a python package instead of FSL? FSL appears to be cumbersome and poorly documented.
I would really appreciate any new inputs.
P.S. I think I cannot use nilearn as its documentation is only for multiple subject ICA.
Thank you
I see that you have asked the same question in fMRI single subject ICA decomposition python based.
Did canICA not work for you?
I am not sure that CanICA can be applied to single-subject ICA. Documentation on CanICA says it is used for multi-subject ICA.
Would it be instead appropriate to loop through the subjects using CanICA on one subject’s fMRI at a time?
Thank you so much
I have not tried CanICA yet. Doing single subject ICA is also on my to-do, which is why I am interested.
I think the person who suggested you can use it for single subjects in fMRI single subject ICA decomposition python based (@GaelVaroquaux) is the author of CanICa (see https://www.sciencedirect.com/science/article/pii/S1053811910001618), so I think he would know.
Perhaps @GaelVaroquaux could chime in and clarify?
Dror
That is great, thank you so very much for the information.
No worries.
You may also find the ICA section in this paper and the accompanying code handy.
Again, its on my todo but have not got around to it yet.
The nilearn code treats the single-subject case as a specific case of the multi-subject (see the code at https://github.com/nilearn/nilearn/blob/master/nilearn/decomposition/base.py#L380 ). In practice, it means that the PCA is performed directly on the subject data: it is a classic single-subject model.
However, if you have multiple sessions, I suggest that you use CanICA on the multiple sessions as you would in a multi-subject setting: the hierarchical model is still of interest.
The documentation could be improved  .
.
That is great, thank you so much for your reply. Another question comes up as I’m learning this: in FSL Melodic software, the program runs single subject ICA and creates both a spatial map and power spectrums per each component, also returning txt files with component frequencies. Is there a way to do the same with nilearn or nitime? In particular, how can you extract time series from components and then create a fourier transform to get power spectrum?
Also, is there a way for nilearn to automatically determine the number of components (like melodic does).
It would be nice not having to rely on FSL for all this- nilearn is much faster!
Thank you so much again,
S
Hello, I also need to use ICA as a single object extraction network. But in the process of using it, I found some differences. An ADHD data and my data are shown below(Both use an object). My data shows that there is something wrong with the results. Is the result normal? Is it my problem of data preprocessing? Or is there a problem with some codes?
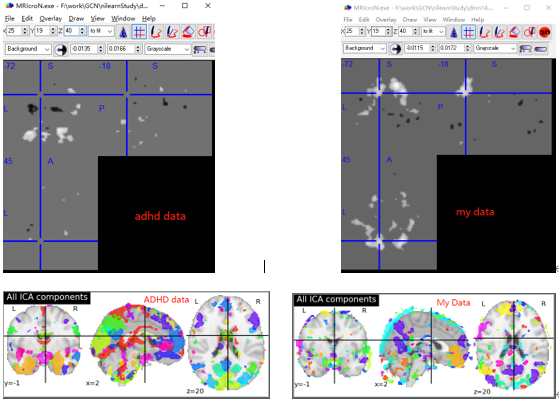
codes:
num = 40
adhd_data = datasets.fetch_adhd(n_subjects=num)
func = adhd_data.func[0]
# func = r'I:\TenDrugDataNoBrain\Drug01_brain.nii' # my data
canica = decomposition.CanICA(n_components=20, mask_strategy='background')
#
# # # func_filenames = rest_dataset.func
canica.fit(func)
#Retrieving the components
components = canica.components_
# canica_components_img = canica.components_img_
#Using a masker to project into the 3D space
components_img = canica.masker_.inverse_transform(components)
canica_components_img = image.index_img(components_img, 9)
#Plotting the default mode network (DMN) without region extraction
plotting.plot_stat_map(image.index_img(components_img, 9), title='DMN')
plotting.show()
# Plotting all the components
plotting.plot_prob_atlas(components_img, title='All ICA components')
plotting.show()
canica_components_img.to_filename('dmn13.nii.gz')