Summary of what happened:
Hi,
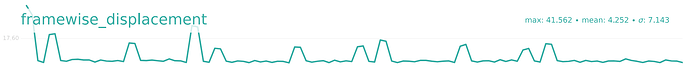
I’m using QSIprep 0.22.0 for preprocessing a dMRI dataset. For two subjects, the preprocessed data seem stripped -but not as the whole slices are missing- evident as early as in the denoising report:
The raw data has no evident issues. I repeated the analysis with QSIprep 1.0.2; the issue remained, but different slices were missing.
Command used (and if a helper script was used, a link to the helper script or the command generated):
Here is my QSIprep call:
singularity run --nv --cleanenv -B $TMPDIR:$TMPDIR \
qsiprep-v0.22.0.sif /input/ /output/derivatives/ participant \
--fs-license-file /toolboxes/license.txt \
--participant-label ${subjid} --output-resolution 1.25 \
--denoise-method dwidenoise --unringing-method mrdegibbs \
--hmc-model eddy --eddy-config /output/code/eddy_params.json --pepolar-method TOPUP \
--anat-modality T1w --anatomical-template MNI152NLin2009cAsym --b0_to_t1w_transform Rigid \
-w /user/jije2119/work --freesurfer-input /output/derivatives/fastsurfer_2.3.0 \
--stop-on-first-crash --nthreads 30 --skip_bids_validation
Here is the eddy_params.json file:
{
"flm": "quadratic",
"slm": "linear",
"fep": false,
"interp": "spline",
"nvoxhp": 1000,
"fudge_factor": 10,
"dont_sep_offs_move": false,
"dont_peas": false,
"niter": 5,
"method": "jac",
"repol": true,
"num_threads": 1,
"is_shelled": true,
"use_cuda": true,
"cnr_maps": true,
"residuals": true,
"output_type": "NIFTI_GZ",
"estimate_move_by_susceptibility": true,
"mporder": 6,
"args": "--ol_nstd=5"
}
Version:
0.22.0 and 1.0.2
Environment (Docker, Singularity / Apptainer, custom installation):
Singularity
Data formatted according to a validatable standard? Please provide the output of the validator:
I did not BIDS-validate the data, but 60 other subjects have been successfully preprocessed.
Relevant log outputs (up to 20 lines):
The preprocessing pipeline finishes normally without any error.
Screenshots / relevant information:
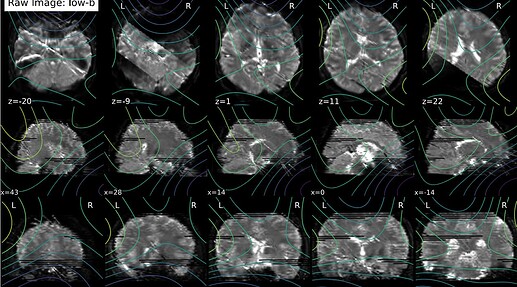
If necessary, I have 75 slices with a multi-band factor of 3. I did some additional debugging by looking in the work directory, and I’m not sure whether this is important, but I noticed that dwi_preproc_wf/hmc_sdc_wf/extract_b0_series/eddy_corrected_LPS_b0_series_mean.nii.gz has cuts (see below), whereas it isn’t in other subjects.
Any comments are appreciated.
Best,
Amir